Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 579
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2754 | 1.488e-02 | 1.166e-03 | 4.016e-01 | 6.969e-06 |
|---|
| IPC | 0.4407 | 1.922e-01 | 6.519e-02 | 9.005e-01 | 1.128e-02 |
|---|
| Loi | 0.4340 | 3.550e-04 | 0.000e+00 | 7.816e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6374 | 0.000e+00 | 0.000e+00 | 5.332e-02 | 0.000e+00 |
|---|
| Wang | 0.2389 | 1.545e-02 | 1.027e-01 | 3.153e-01 | 5.004e-04 |
|---|
Expression data for subnetwork 579 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
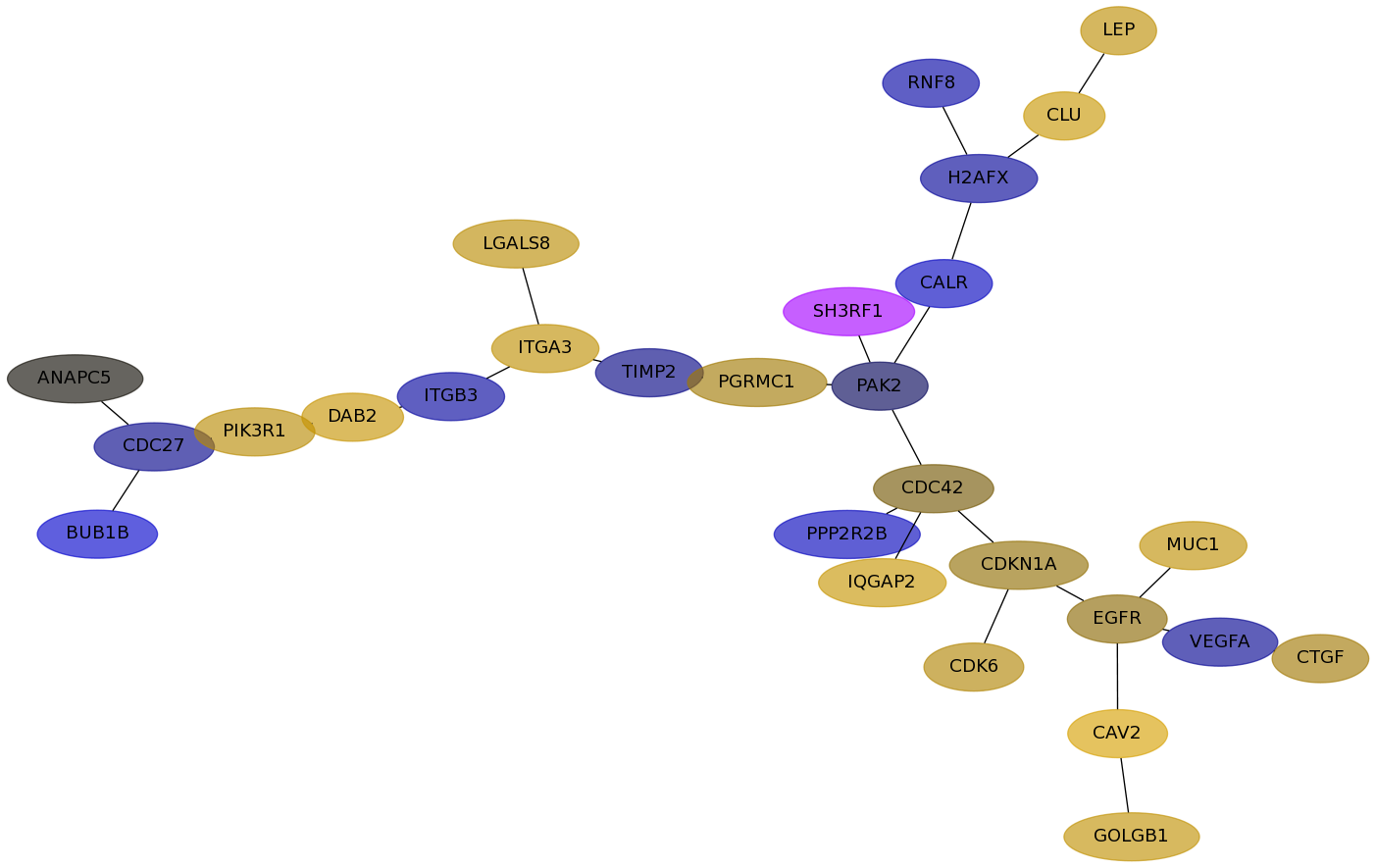
Score for each gene in subnetwork 579 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| ANAPC5 |   | 1 | 652 | 117 | 169 | 0.002 | -0.190 | 0.206 | -0.074 | 0.154 |
|---|
| TIMP2 |   | 3 | 265 | 1 | 24 | -0.053 | -0.126 | 0.212 | 0.021 | 0.137 |
|---|
| LGALS8 |   | 2 | 391 | 1 | 27 | 0.197 | 0.081 | 0.002 | -0.057 | -0.071 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| BUB1B |   | 2 | 391 | 57 | 80 | -0.289 | 0.050 | -0.164 | 0.037 | 0.148 |
|---|
| RNF8 |   | 2 | 391 | 117 | 135 | -0.115 | 0.216 | 0.004 | 0.114 | 0.221 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| CALR |   | 10 | 67 | 117 | 121 | -0.216 | -0.105 | 0.002 | -0.061 | -0.075 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| CDC27 |   | 4 | 198 | 1 | 23 | -0.062 | 0.156 | -0.086 | -0.029 | 0.348 |
|---|
| ITGA3 |   | 3 | 265 | 1 | 24 | 0.218 | -0.135 | 0.299 | 0.037 | -0.104 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| ITGB3 |   | 3 | 265 | 1 | 24 | -0.100 | -0.015 | 0.030 | 0.086 | 0.085 |
|---|
| PGRMC1 |   | 1 | 652 | 117 | 169 | 0.109 | -0.161 | 0.036 | 0.044 | -0.007 |
|---|
| H2AFX |   | 4 | 198 | 117 | 125 | -0.086 | 0.111 | -0.072 | 0.121 | -0.010 |
|---|
| LEP |   | 2 | 391 | 117 | 135 | 0.212 | -0.014 | -0.019 | 0.145 | 0.059 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| GOLGB1 |   | 3 | 265 | 117 | 127 | 0.231 | 0.104 | 0.352 | 0.044 | 0.175 |
|---|
GO Enrichment output for subnetwork 579 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 6.113E-07 | 1.406E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.113E-07 | 7.03E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.219E-06 | 9.346E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.127E-06 | 1.223E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.127E-06 | 9.785E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 2.902E-06 | 1.113E-03 |
|---|
| nucleus localization | GO:0051647 |  | 5.076E-06 | 1.668E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 7.231E-06 | 2.079E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 9.914E-06 | 2.534E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.318E-05 | 3.032E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.411E-05 | 5.042E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 4.166E-07 | 1.018E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.166E-07 | 5.089E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.162E-06 | 9.461E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.162E-06 | 7.096E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 1.182E-06 | 5.777E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.739E-06 | 7.082E-04 |
|---|
| regulation of MAPKKK cascade | GO:0043408 |  | 1.827E-06 | 6.375E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.403E-06 | 1.039E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 4.528E-06 | 1.229E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.373E-06 | 2.046E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.385E-05 | 3.077E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 5.48E-07 | 1.319E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.48E-07 | 6.593E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.57E-07 | 7.675E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 9.57E-07 | 5.756E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 1.456E-06 | 7.008E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.528E-06 | 6.126E-04 |
|---|
| nucleus localization | GO:0051647 |  | 4.472E-06 | 1.537E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.949E-06 | 1.789E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.112E-05 | 2.972E-03 |
|---|
| interphase | GO:0051325 |  | 1.294E-05 | 3.112E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.501E-05 | 3.283E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 6.113E-07 | 1.406E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.113E-07 | 7.03E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.219E-06 | 9.346E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.127E-06 | 1.223E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.127E-06 | 9.785E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 2.902E-06 | 1.113E-03 |
|---|
| nucleus localization | GO:0051647 |  | 5.076E-06 | 1.668E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 7.231E-06 | 2.079E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 9.914E-06 | 2.534E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.318E-05 | 3.032E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.411E-05 | 5.042E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 6.113E-07 | 1.406E-03 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.113E-07 | 7.03E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.219E-06 | 9.346E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.127E-06 | 1.223E-03 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.127E-06 | 9.785E-04 |
|---|
| regulation of lipid metabolic process | GO:0019216 |  | 2.902E-06 | 1.113E-03 |
|---|
| nucleus localization | GO:0051647 |  | 5.076E-06 | 1.668E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 7.231E-06 | 2.079E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 9.914E-06 | 2.534E-03 |
|---|
| microspike assembly | GO:0030035 |  | 1.318E-05 | 3.032E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.411E-05 | 5.042E-03 |
|---|


