Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 565
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2783 | 1.347e-02 | 9.870e-04 | 3.878e-01 | 5.154e-06 |
|---|
| IPC | 0.4408 | 1.922e-01 | 6.516e-02 | 9.005e-01 | 1.128e-02 |
|---|
| Loi | 0.4325 | 3.780e-04 | 0.000e+00 | 7.920e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6323 | 0.000e+00 | 0.000e+00 | 5.956e-02 | 0.000e+00 |
|---|
| Wang | 0.2349 | 1.780e-02 | 1.120e-01 | 3.352e-01 | 6.685e-04 |
|---|
Expression data for subnetwork 565 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
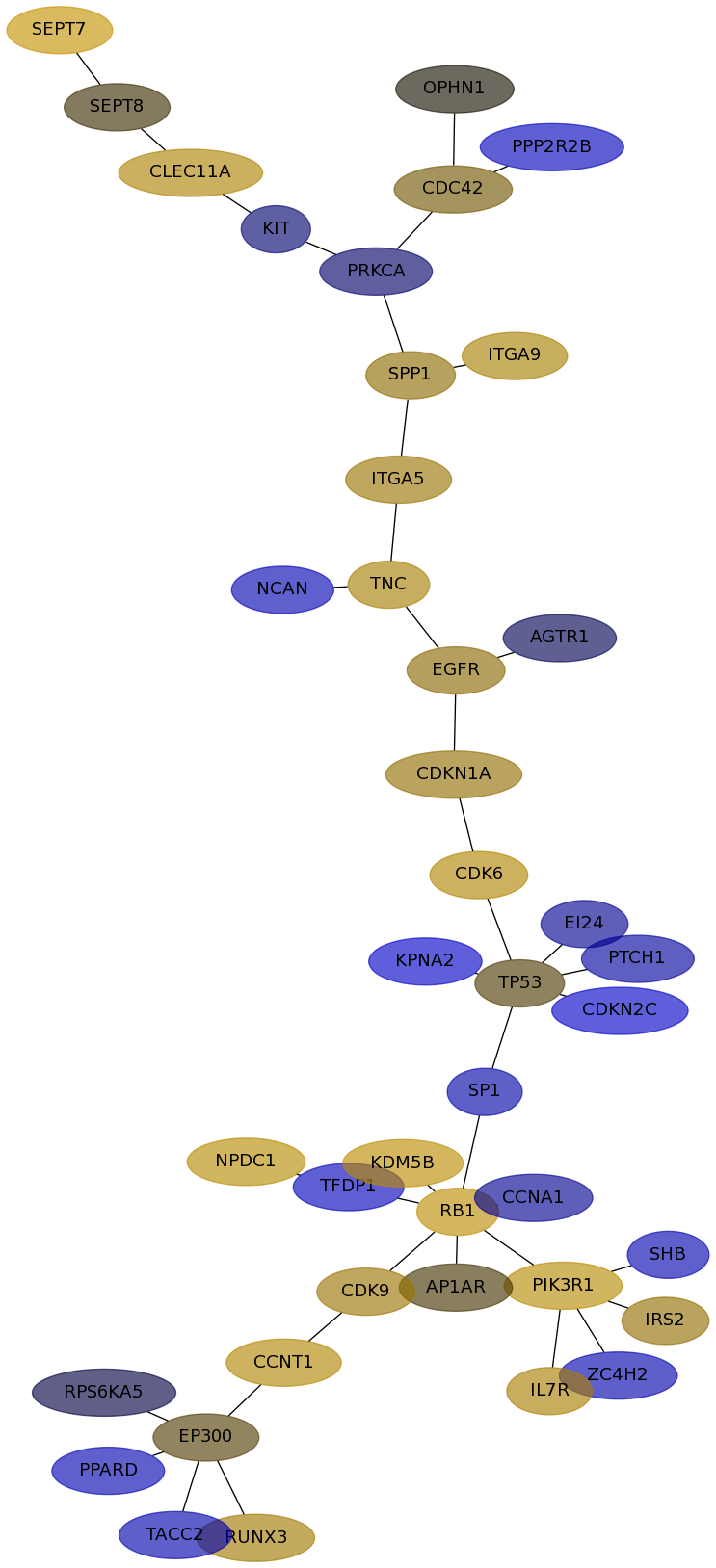
Score for each gene in subnetwork 565 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| SHB |   | 2 | 391 | 339 | 352 | -0.160 | 0.164 | 0.141 | -0.074 | -0.048 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| OPHN1 |   | 5 | 148 | 339 | 337 | 0.003 | 0.062 | 0.278 | 0.194 | -0.182 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| PRKCA |   | 5 | 148 | 339 | 337 | -0.029 | -0.016 | -0.088 | 0.174 | 0.005 |
|---|
| RPS6KA5 |   | 1 | 652 | 339 | 366 | -0.011 | 0.024 | -0.081 | 0.118 | -0.161 |
|---|
| TNC |   | 2 | 391 | 339 | 352 | 0.125 | -0.154 | 0.282 | 0.113 | 0.072 |
|---|
| CDKN2C |   | 3 | 265 | 283 | 292 | -0.269 | 0.216 | -0.106 | 0.202 | -0.081 |
|---|
| SPP1 |   | 4 | 198 | 339 | 341 | 0.068 | 0.130 | 0.119 | 0.184 | 0.153 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| TACC2 |   | 9 | 77 | 148 | 154 | -0.151 | -0.073 | 0.122 | 0.181 | 0.180 |
|---|
| EI24 |   | 8 | 84 | 339 | 336 | -0.074 | 0.051 | 0.078 | 0.107 | 0.070 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| ITGA9 |   | 1 | 652 | 339 | 366 | 0.132 | -0.180 | 0.089 | 0.145 | 0.036 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| CLEC11A |   | 5 | 148 | 339 | 337 | 0.146 | 0.004 | 0.353 | 0.005 | 0.272 |
|---|
| KDM5B |   | 3 | 265 | 339 | 348 | 0.199 | -0.011 | 0.279 | -0.047 | 0.161 |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PTCH1 |   | 4 | 198 | 339 | 341 | -0.102 | -0.184 | 0.047 | 0.121 | 0.004 |
|---|
| SEPT8 |   | 2 | 391 | 339 | 352 | 0.010 | 0.209 | 0.148 | 0.097 | 0.169 |
|---|
| ITGA5 |   | 5 | 148 | 339 | 337 | 0.093 | 0.031 | 0.192 | 0.125 | 0.202 |
|---|
| RUNX3 |   | 8 | 84 | 33 | 35 | 0.109 | 0.071 | -0.097 | 0.083 | -0.074 |
|---|
| NCAN |   | 1 | 652 | 339 | 366 | -0.161 | -0.035 | -0.058 | 0.102 | -0.047 |
|---|
| SEPT7 |   | 1 | 652 | 339 | 366 | 0.241 | 0.224 | 0.025 | -0.016 | 0.019 |
|---|
| NPDC1 |   | 3 | 265 | 339 | 348 | 0.190 | -0.019 | 0.221 | 0.044 | -0.146 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| IRS2 |   | 1 | 652 | 339 | 366 | 0.076 | -0.006 | 0.043 | 0.054 | 0.125 |
|---|
| PPARD |   | 1 | 652 | 339 | 366 | -0.158 | -0.006 | 0.091 | -0.002 | 0.194 |
|---|
| AP1AR |   | 1 | 652 | 339 | 366 | 0.012 | 0.059 | 0.004 | 0.146 | -0.011 |
|---|
| AGTR1 |   | 1 | 652 | 339 | 366 | -0.017 | 0.025 | -0.058 | 0.174 | -0.007 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| CDK9 |   | 4 | 198 | 129 | 130 | 0.095 | -0.120 | 0.179 | 0.182 | -0.135 |
|---|
| TFDP1 |   | 4 | 198 | 339 | 341 | -0.194 | 0.123 | -0.178 | 0.129 | -0.065 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| KPNA2 |   | 1 | 652 | 339 | 366 | -0.266 | 0.233 | -0.081 | 0.087 | -0.090 |
|---|
GO Enrichment output for subnetwork 565 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.169E-09 | 2.109E-05 |
|---|
| negative regulation of growth | GO:0045926 |  | 2.369E-08 | 2.724E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.268E-08 | 2.506E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.127E-08 | 2.948E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.186E-08 | 2.846E-05 |
|---|
| interphase | GO:0051325 |  | 7.262E-08 | 2.784E-05 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 2.022E-07 | 6.644E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 2.45E-07 | 7.045E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 5.313E-07 | 1.358E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.053E-07 | 1.392E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 1.392E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.712E-09 | 1.64E-05 |
|---|
| negative regulation of growth | GO:0045926 |  | 1.094E-08 | 1.336E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.203E-08 | 9.796E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.852E-08 | 1.131E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.964E-08 | 1.448E-05 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 7.694E-08 | 3.133E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 9.195E-08 | 3.209E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.279E-07 | 1.001E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.279E-07 | 8.9E-05 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 3.618E-07 | 8.838E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.669E-07 | 8.149E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.974E-09 | 9.561E-06 |
|---|
| negative regulation of growth | GO:0045926 |  | 1.321E-08 | 1.589E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.475E-08 | 1.183E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.27E-08 | 1.366E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.129E-08 | 1.506E-05 |
|---|
| interphase | GO:0051325 |  | 3.886E-08 | 1.558E-05 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 9.754E-08 | 3.352E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 1.165E-07 | 3.505E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 3.177E-07 | 8.493E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.011E-07 | 9.651E-05 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 4.268E-07 | 9.335E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.169E-09 | 2.109E-05 |
|---|
| negative regulation of growth | GO:0045926 |  | 2.369E-08 | 2.724E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.268E-08 | 2.506E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.127E-08 | 2.948E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.186E-08 | 2.846E-05 |
|---|
| interphase | GO:0051325 |  | 7.262E-08 | 2.784E-05 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 2.022E-07 | 6.644E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 2.45E-07 | 7.045E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 5.313E-07 | 1.358E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.053E-07 | 1.392E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 1.392E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.169E-09 | 2.109E-05 |
|---|
| negative regulation of growth | GO:0045926 |  | 2.369E-08 | 2.724E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.268E-08 | 2.506E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.127E-08 | 2.948E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.186E-08 | 2.846E-05 |
|---|
| interphase | GO:0051325 |  | 7.262E-08 | 2.784E-05 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 2.022E-07 | 6.644E-05 |
|---|
| negative regulation of cell size | GO:0045792 |  | 2.45E-07 | 7.045E-05 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 5.313E-07 | 1.358E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.053E-07 | 1.392E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.659E-07 | 1.392E-04 |
|---|


