Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 524
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2813 | 1.213e-02 | 8.290e-04 | 3.737e-01 | 3.758e-06 |
|---|
| IPC | 0.4481 | 1.821e-01 | 5.955e-02 | 8.980e-01 | 9.737e-03 |
|---|
| Loi | 0.4229 | 5.580e-04 | 0.000e+00 | 8.601e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6174 | 0.000e+00 | 0.000e+00 | 8.104e-02 | 0.000e+00 |
|---|
| Wang | 0.2316 | 1.998e-02 | 1.203e-01 | 3.521e-01 | 8.462e-04 |
|---|
Expression data for subnetwork 524 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
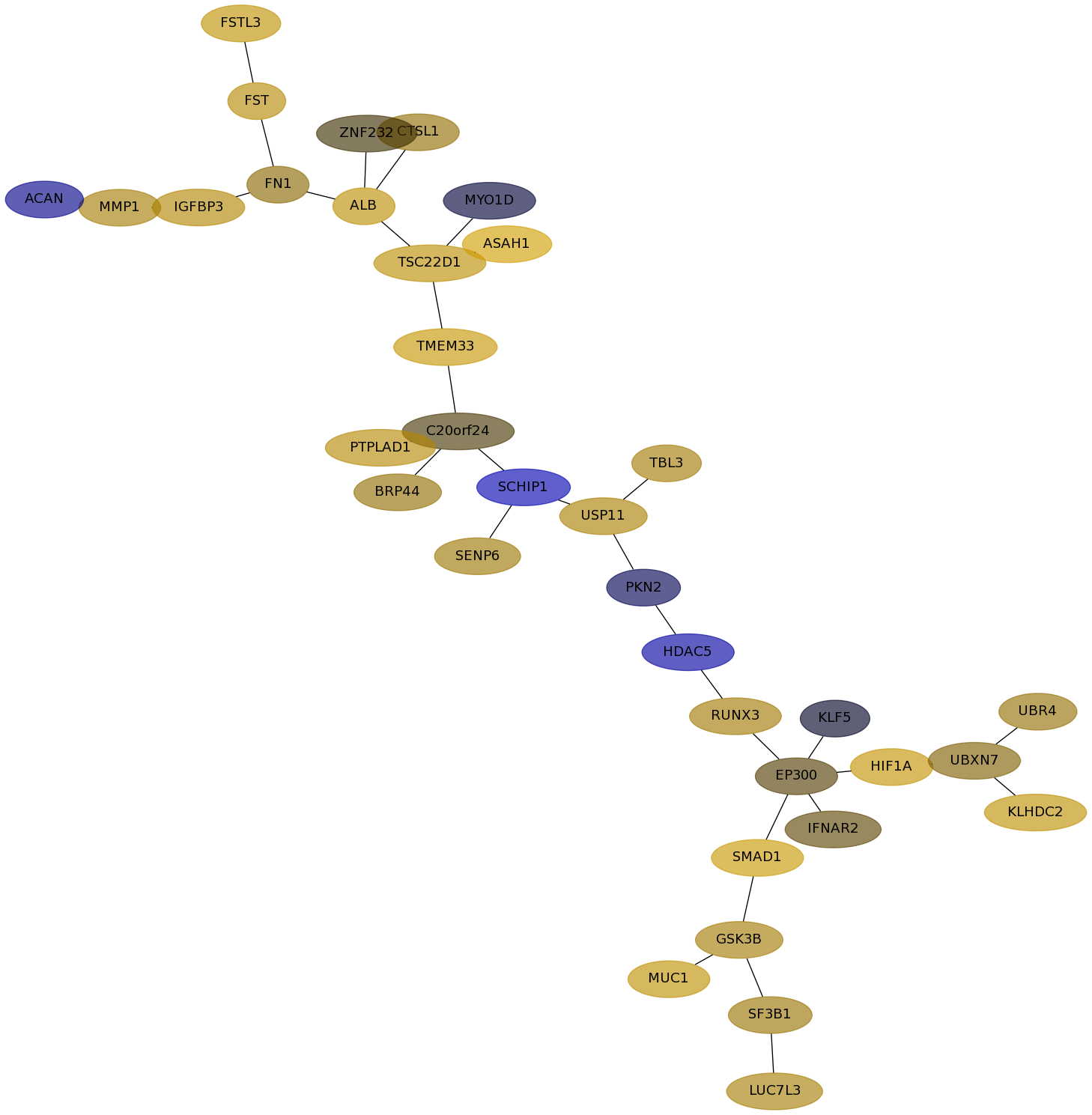
Score for each gene in subnetwork 524 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| MYO1D |   | 2 | 391 | 563 | 567 | -0.008 | 0.052 | 0.265 | -0.154 | 0.013 |
|---|
| IFNAR2 |   | 4 | 198 | 33 | 41 | 0.021 | -0.100 | 0.055 | 0.106 | -0.058 |
|---|
| FN1 |   | 5 | 148 | 57 | 63 | 0.064 | 0.190 | 0.133 | 0.091 | 0.083 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| ASAH1 |   | 1 | 652 | 1039 | 1048 | 0.343 | -0.170 | -0.018 | -0.105 | 0.002 |
|---|
| PKN2 |   | 3 | 265 | 552 | 550 | -0.017 | -0.198 | 0.158 | 0.060 | 0.068 |
|---|
| UBXN7 |   | 6 | 120 | 148 | 156 | 0.049 | -0.055 | 0.153 | 0.224 | 0.175 |
|---|
| C20orf24 |   | 1 | 652 | 1039 | 1048 | 0.013 | 0.041 | -0.145 | -0.184 | 0.010 |
|---|
| SCHIP1 |   | 2 | 391 | 1039 | 1033 | -0.160 | 0.130 | 0.136 | 0.105 | 0.198 |
|---|
| TSC22D1 |   | 1 | 652 | 1039 | 1048 | 0.206 | -0.196 | 0.158 | 0.046 | 0.011 |
|---|
| SENP6 |   | 1 | 652 | 1039 | 1048 | 0.099 | 0.037 | 0.122 | 0.135 | 0.125 |
|---|
| IGFBP3 |   | 2 | 391 | 1039 | 1033 | 0.165 | -0.033 | -0.031 | 0.034 | -0.065 |
|---|
| KLHDC2 |   | 2 | 391 | 923 | 915 | 0.219 | 0.082 | 0.065 | -0.030 | 0.079 |
|---|
| MMP1 |   | 3 | 265 | 1039 | 1013 | 0.122 | 0.192 | 0.014 | 0.036 | 0.260 |
|---|
| RUNX3 |   | 8 | 84 | 33 | 35 | 0.109 | 0.071 | -0.097 | 0.083 | -0.074 |
|---|
| UBR4 |   | 2 | 391 | 1039 | 1033 | 0.079 | -0.093 | 0.268 | -0.079 | 0.061 |
|---|
| HDAC5 |   | 2 | 391 | 33 | 55 | -0.107 | 0.022 | 0.029 | -0.165 | 0.009 |
|---|
| CTSL1 |   | 3 | 265 | 283 | 292 | 0.076 | 0.187 | -0.080 | 0.091 | 0.178 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| ZNF232 |   | 5 | 148 | 57 | 63 | 0.010 | 0.048 | -0.040 | 0.189 | 0.159 |
|---|
| BRP44 |   | 1 | 652 | 1039 | 1048 | 0.079 | -0.047 | 0.075 | -0.069 | 0.067 |
|---|
| ALB |   | 7 | 100 | 57 | 60 | 0.207 | -0.122 | 0.218 | 0.080 | 0.038 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| TBL3 |   | 1 | 652 | 1039 | 1048 | 0.113 | 0.074 | -0.067 | -0.033 | -0.031 |
|---|
| ACAN |   | 4 | 198 | 797 | 792 | -0.060 | 0.156 | 0.132 | -0.024 | -0.083 |
|---|
| FSTL3 |   | 1 | 652 | 1039 | 1048 | 0.228 | 0.051 | 0.155 | 0.001 | 0.003 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| PTPLAD1 |   | 1 | 652 | 1039 | 1048 | 0.175 | -0.183 | 0.036 | -0.078 | -0.100 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| FST |   | 1 | 652 | 1039 | 1048 | 0.183 | 0.036 | 0.298 | 0.010 | -0.021 |
|---|
| USP11 |   | 5 | 148 | 283 | 287 | 0.134 | -0.144 | 0.106 | 0.121 | -0.044 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| TMEM33 |   | 3 | 265 | 1030 | 1008 | 0.264 | -0.074 | -0.077 | -0.187 | 0.033 |
|---|
GO Enrichment output for subnetwork 524 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 1.112E-04 | 0.25586782 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 1.112E-04 | 0.12793391 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.112E-04 | 0.08528927 |
|---|
| microvillus organization | GO:0032528 |  | 1.112E-04 | 0.06396695 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 1.112E-04 | 0.05117356 |
|---|
| regulation of hair follicle development | GO:0051797 |  | 1.112E-04 | 0.04264464 |
|---|
| BMP signaling pathway | GO:0030509 |  | 1.126E-04 | 0.03701227 |
|---|
| collagen metabolic process | GO:0032963 |  | 1.389E-04 | 0.03994408 |
|---|
| regulation of striated muscle tissue development | GO:0016202 |  | 1.535E-04 | 0.03921589 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.665E-04 | 0.0382968 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 1.665E-04 | 0.03481527 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 6.169E-05 | 0.15071215 |
|---|
| positive regulation of muscle cell differentiation | GO:0051149 |  | 6.169E-05 | 0.07535608 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 6.169E-05 | 0.05023738 |
|---|
| microvillus organization | GO:0032528 |  | 6.169E-05 | 0.03767804 |
|---|
| mRNA transcription | GO:0009299 |  | 6.169E-05 | 0.03014243 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 6.169E-05 | 0.02511869 |
|---|
| regulation of hair follicle development | GO:0051797 |  | 6.169E-05 | 0.02153031 |
|---|
| collagen metabolic process | GO:0032963 |  | 7.141E-05 | 0.02180717 |
|---|
| BMP signaling pathway | GO:0030509 |  | 7.141E-05 | 0.01938415 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 9.239E-05 | 0.02256995 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 9.239E-05 | 0.02051814 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| hemoglobin biosynthetic process | GO:0042541 |  | 7.464E-05 | 0.17957286 |
|---|
| positive regulation of muscle cell differentiation | GO:0051149 |  | 7.464E-05 | 0.08978643 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 7.464E-05 | 0.05985762 |
|---|
| microvillus organization | GO:0032528 |  | 7.464E-05 | 0.04489322 |
|---|
| mRNA transcription | GO:0009299 |  | 7.464E-05 | 0.03591457 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 7.464E-05 | 0.02992881 |
|---|
| regulation of hair follicle development | GO:0051797 |  | 7.464E-05 | 0.02565327 |
|---|
| collagen metabolic process | GO:0032963 |  | 8.58E-05 | 0.02580495 |
|---|
| BMP signaling pathway | GO:0030509 |  | 9.449E-05 | 0.02526032 |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 1.118E-04 | 0.02688766 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 1.118E-04 | 0.02444333 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 1.112E-04 | 0.25586782 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 1.112E-04 | 0.12793391 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.112E-04 | 0.08528927 |
|---|
| microvillus organization | GO:0032528 |  | 1.112E-04 | 0.06396695 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 1.112E-04 | 0.05117356 |
|---|
| regulation of hair follicle development | GO:0051797 |  | 1.112E-04 | 0.04264464 |
|---|
| BMP signaling pathway | GO:0030509 |  | 1.126E-04 | 0.03701227 |
|---|
| collagen metabolic process | GO:0032963 |  | 1.389E-04 | 0.03994408 |
|---|
| regulation of striated muscle tissue development | GO:0016202 |  | 1.535E-04 | 0.03921589 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.665E-04 | 0.0382968 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 1.665E-04 | 0.03481527 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of hormone biosynthetic process | GO:0046885 |  | 1.112E-04 | 0.25586782 |
|---|
| embryonic hemopoiesis | GO:0035162 |  | 1.112E-04 | 0.12793391 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.112E-04 | 0.08528927 |
|---|
| microvillus organization | GO:0032528 |  | 1.112E-04 | 0.06396695 |
|---|
| positive regulation of chemokine production | GO:0032722 |  | 1.112E-04 | 0.05117356 |
|---|
| regulation of hair follicle development | GO:0051797 |  | 1.112E-04 | 0.04264464 |
|---|
| BMP signaling pathway | GO:0030509 |  | 1.126E-04 | 0.03701227 |
|---|
| collagen metabolic process | GO:0032963 |  | 1.389E-04 | 0.03994408 |
|---|
| regulation of striated muscle tissue development | GO:0016202 |  | 1.535E-04 | 0.03921589 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.665E-04 | 0.0382968 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 1.665E-04 | 0.03481527 |
|---|


