Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 301
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.2823 | 1.172e-02 | 7.830e-04 | 3.691e-01 | 3.388e-06 |
|---|
| IPC | 0.4108 | 2.362e-01 | 9.210e-02 | 9.111e-01 | 1.982e-02 |
|---|
| Loi | 0.3950 | 1.613e-03 | 0.000e+00 | 1.091e-01 | 0.000e+00 |
|---|
| Schmidt | 0.5414 | 1.800e-05 | 0.000e+00 | 2.772e-01 | 0.000e+00 |
|---|
| Wang | 0.1718 | 1.063e-01 | 3.272e-01 | 6.648e-01 | 2.312e-02 |
|---|
Expression data for subnetwork 301 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
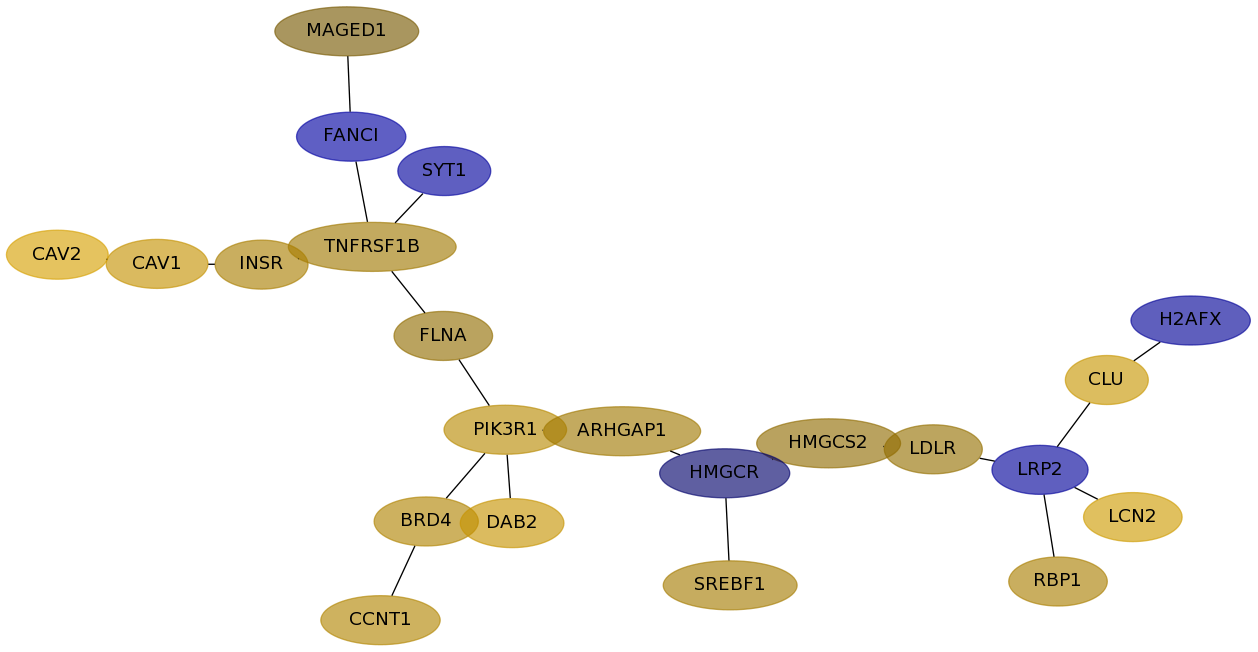
Score for each gene in subnetwork 301 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| LCN2 |   | 6 | 120 | 797 | 779 | 0.316 | 0.113 | 0.145 | -0.098 | -0.284 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| LDLR |   | 3 | 265 | 679 | 667 | 0.085 | -0.041 | 0.218 | 0.039 | 0.064 |
|---|
| MAGED1 |   | 6 | 120 | 715 | 695 | 0.040 | -0.141 | 0.284 | -0.152 | 0.045 |
|---|
| HMGCS2 |   | 3 | 265 | 129 | 139 | 0.073 | -0.277 | 0.226 | -0.194 | -0.085 |
|---|
| LRP2 |   | 2 | 391 | 1177 | 1166 | -0.092 | 0.193 | -0.042 | 0.001 | 0.125 |
|---|
| FLNA |   | 7 | 100 | 206 | 198 | 0.077 | 0.081 | 0.107 | 0.148 | 0.126 |
|---|
| CAV1 |   | 10 | 67 | 439 | 428 | 0.251 | -0.092 | 0.124 | 0.079 | 0.119 |
|---|
| SYT1 |   | 1 | 652 | 1481 | 1481 | -0.102 | -0.079 | 0.051 | 0.038 | -0.124 |
|---|
| FANCI |   | 4 | 198 | 832 | 809 | -0.111 | 0.145 | -0.194 | 0.104 | -0.036 |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| H2AFX |   | 4 | 198 | 117 | 125 | -0.086 | 0.111 | -0.072 | 0.121 | -0.010 |
|---|
| SREBF1 |   | 2 | 391 | 859 | 860 | 0.119 | -0.042 | 0.232 | -0.195 | -0.074 |
|---|
| ARHGAP1 |   | 3 | 265 | 129 | 139 | 0.108 | -0.167 | 0.297 | 0.001 | 0.251 |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| RBP1 |   | 2 | 391 | 1177 | 1166 | 0.133 | -0.002 | -0.005 | 0.112 | -0.089 |
|---|
| HMGCR |   | 3 | 265 | 129 | 139 | -0.029 | -0.075 | 0.226 | 0.026 | -0.016 |
|---|
GO Enrichment output for subnetwork 301 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.482E-07 | 3.409E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.482E-07 | 1.705E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.642E-07 | 1.259E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 2.959E-07 | 1.702E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.959E-07 | 1.361E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.959E-07 | 1.134E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.17E-07 | 1.699E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 5.899E-07 | 1.696E-04 |
|---|
| cholesterol homeostasis | GO:0042632 |  | 6.837E-07 | 1.747E-04 |
|---|
| negative regulation of MAP kinase activity | GO:0043407 |  | 6.837E-07 | 1.573E-04 |
|---|
| cholesterol transport | GO:0030301 |  | 9.118E-07 | 1.906E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cholesterol metabolic process | GO:0008203 |  | 6.363E-09 | 1.554E-05 |
|---|
| sterol metabolic process | GO:0016125 |  | 1.171E-08 | 1.43E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.011E-08 | 3.266E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.334E-08 | 2.647E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 1.513E-07 | 7.394E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.513E-07 | 6.161E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 2.174E-07 | 7.586E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 2.418E-07 | 7.385E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.418E-07 | 6.564E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.418E-07 | 5.908E-05 |
|---|
| cholesterol homeostasis | GO:0042632 |  | 2.702E-07 | 6.001E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.079E-08 | 1.222E-04 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 6.073E-08 | 7.306E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.213E-07 | 9.728E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.12E-07 | 1.275E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.12E-07 | 1.02E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.12E-07 | 8.501E-05 |
|---|
| cholesterol homeostasis | GO:0042632 |  | 3.249E-07 | 1.117E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 3.524E-07 | 1.06E-04 |
|---|
| cholesterol transport | GO:0030301 |  | 3.707E-07 | 9.911E-05 |
|---|
| negative regulation of MAP kinase activity | GO:0043407 |  | 3.707E-07 | 8.92E-05 |
|---|
| sterol transport | GO:0015918 |  | 4.213E-07 | 9.215E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.482E-07 | 3.409E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.482E-07 | 1.705E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.642E-07 | 1.259E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 2.959E-07 | 1.702E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.959E-07 | 1.361E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.959E-07 | 1.134E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.17E-07 | 1.699E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 5.899E-07 | 1.696E-04 |
|---|
| cholesterol homeostasis | GO:0042632 |  | 6.837E-07 | 1.747E-04 |
|---|
| negative regulation of MAP kinase activity | GO:0043407 |  | 6.837E-07 | 1.573E-04 |
|---|
| cholesterol transport | GO:0030301 |  | 9.118E-07 | 1.906E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.482E-07 | 3.409E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.482E-07 | 1.705E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.642E-07 | 1.259E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 2.959E-07 | 1.702E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.959E-07 | 1.361E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.959E-07 | 1.134E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.17E-07 | 1.699E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 5.899E-07 | 1.696E-04 |
|---|
| cholesterol homeostasis | GO:0042632 |  | 6.837E-07 | 1.747E-04 |
|---|
| negative regulation of MAP kinase activity | GO:0043407 |  | 6.837E-07 | 1.573E-04 |
|---|
| cholesterol transport | GO:0030301 |  | 9.118E-07 | 1.906E-04 |
|---|


