Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1292
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3377 | 1.236e-03 | 1.900e-05 | 1.490e-01 | 3.498e-09 |
|---|
| IPC | 0.3133 | 3.938e-01 | 2.197e-01 | 9.448e-01 | 8.174e-02 |
|---|
| Loi | 0.4903 | 2.800e-05 | 0.000e+00 | 4.668e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5946 | 1.000e-06 | 0.000e+00 | 1.243e-01 | 0.000e+00 |
|---|
| Wang | 0.2533 | 8.982e-03 | 7.334e-02 | 2.476e-01 | 1.631e-04 |
|---|
Expression data for subnetwork 1292 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
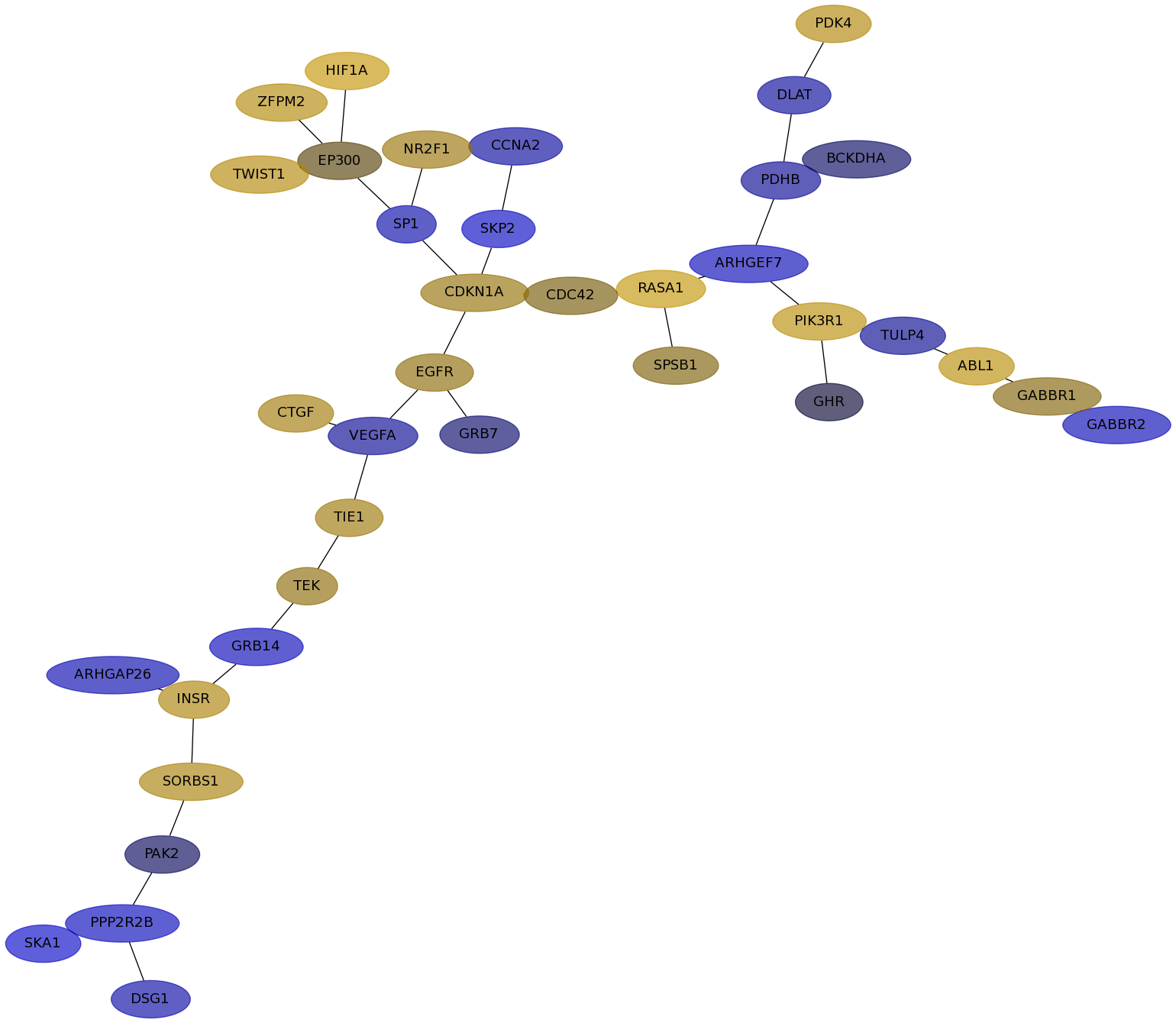
Score for each gene in subnetwork 1292 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| TEK |   | 7 | 100 | 148 | 155 | 0.063 | -0.149 | -0.049 | 0.174 | 0.113 |
|---|
| ARHGAP26 |   | 5 | 148 | 444 | 440 | -0.145 | -0.023 | 0.171 | -0.222 | -0.348 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| GRB14 |   | 4 | 198 | 977 | 965 | -0.189 | 0.113 | -0.129 | 0.143 | 0.138 |
|---|
| BCKDHA |   | 1 | 652 | 1225 | 1228 | -0.023 | 0.091 | -0.045 | 0.072 | 0.101 |
|---|
| ARHGEF7 |   | 12 | 55 | 492 | 475 | -0.180 | -0.074 | -0.061 | 0.208 | 0.015 |
|---|
| GABBR1 |   | 2 | 391 | 847 | 841 | 0.049 | -0.107 | 0.164 | 0.081 | 0.131 |
|---|
| PDK4 |   | 1 | 652 | 1225 | 1228 | 0.150 | -0.027 | -0.030 | 0.216 | -0.041 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| SPSB1 |   | 7 | 100 | 492 | 495 | 0.044 | -0.124 | 0.088 | -0.152 | 0.169 |
|---|
| ABL1 |   | 8 | 84 | 326 | 316 | 0.193 | -0.152 | 0.200 | 0.108 | -0.059 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| TWIST1 |   | 3 | 265 | 444 | 447 | 0.166 | 0.044 | 0.194 | 0.061 | 0.219 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| TULP4 |   | 1 | 652 | 1225 | 1228 | -0.068 | -0.114 | 0.067 | 0.024 | 0.047 |
|---|
| GABBR2 |   | 1 | 652 | 1225 | 1228 | -0.173 | 0.130 | 0.180 | 0.117 | 0.037 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| ZFPM2 |   | 4 | 198 | 148 | 162 | 0.160 | 0.001 | 0.092 | 0.113 | 0.379 |
|---|
| SKA1 |   | 1 | 652 | 1225 | 1228 | -0.269 | 0.081 | -0.085 | 0.037 | -0.070 |
|---|
| PDHB |   | 1 | 652 | 1225 | 1228 | -0.075 | 0.056 | 0.017 | 0.197 | 0.193 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| NR2F1 |   | 6 | 120 | 444 | 438 | 0.086 | -0.252 | 0.113 | 0.133 | 0.207 |
|---|
| RASA1 |   | 9 | 77 | 492 | 494 | 0.237 | -0.079 | 0.146 | 0.127 | 0.149 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| GRB7 |   | 13 | 47 | 57 | 51 | -0.028 | 0.021 | 0.115 | -0.072 | 0.129 |
|---|
| TIE1 |   | 6 | 120 | 148 | 156 | 0.094 | -0.108 | 0.005 | 0.141 | 0.252 |
|---|
| DSG1 |   | 4 | 198 | 182 | 183 | -0.120 | 0.072 | -0.075 | 0.252 | 0.068 |
|---|
| DLAT |   | 1 | 652 | 1225 | 1228 | -0.089 | 0.111 | 0.029 | 0.072 | -0.014 |
|---|
GO Enrichment output for subnetwork 1292 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.112E-07 | 1.176E-03 |
|---|
| nuclear migration | GO:0007097 |  | 5.112E-07 | 5.879E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.112E-07 | 3.919E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.02E-06 | 5.863E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.02E-06 | 4.69E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.35E-06 | 5.176E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.353E-06 | 4.446E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.554E-06 | 4.468E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.78E-06 | 4.548E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.78E-06 | 4.093E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.86E-06 | 3.889E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| branched chain family amino acid metabolic process | GO:0009081 |  | 1.931E-07 | 4.717E-04 |
|---|
| response to nutrient | GO:0007584 |  | 2.422E-07 | 2.959E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.793E-07 | 2.274E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 4.81E-07 | 2.938E-04 |
|---|
| nuclear migration | GO:0007097 |  | 5.573E-07 | 2.723E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.573E-07 | 2.269E-04 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 8.48E-07 | 2.959E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 9.953E-07 | 3.039E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.187E-06 | 3.223E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.391E-06 | 3.397E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.553E-06 | 3.45E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| branched chain family amino acid metabolic process | GO:0009081 |  | 2.142E-07 | 5.154E-04 |
|---|
| response to nutrient | GO:0007584 |  | 2.423E-07 | 2.915E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 3.027E-07 | 2.427E-04 |
|---|
| gamma-aminobutyric acid signaling pathway | GO:0007214 |  | 4.33E-07 | 2.604E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.039E-07 | 2.906E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.039E-07 | 2.422E-04 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 9.404E-07 | 3.232E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.033E-06 | 3.105E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.054E-06 | 2.819E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.054E-06 | 2.537E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.317E-06 | 2.88E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.112E-07 | 1.176E-03 |
|---|
| nuclear migration | GO:0007097 |  | 5.112E-07 | 5.879E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.112E-07 | 3.919E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.02E-06 | 5.863E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.02E-06 | 4.69E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.35E-06 | 5.176E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.353E-06 | 4.446E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.554E-06 | 4.468E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.78E-06 | 4.548E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.78E-06 | 4.093E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.86E-06 | 3.889E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.112E-07 | 1.176E-03 |
|---|
| nuclear migration | GO:0007097 |  | 5.112E-07 | 5.879E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.112E-07 | 3.919E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.02E-06 | 5.863E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.02E-06 | 4.69E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.35E-06 | 5.176E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.353E-06 | 4.446E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.554E-06 | 4.468E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.78E-06 | 4.548E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.78E-06 | 4.093E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.86E-06 | 3.889E-04 |
|---|


