Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1286
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3381 | 1.214e-03 | 1.800e-05 | 1.478e-01 | 3.229e-09 |
|---|
| IPC | 0.3147 | 3.915e-01 | 2.175e-01 | 9.444e-01 | 8.044e-02 |
|---|
| Loi | 0.4899 | 2.900e-05 | 0.000e+00 | 4.689e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5941 | 1.000e-06 | 0.000e+00 | 1.253e-01 | 0.000e+00 |
|---|
| Wang | 0.2530 | 9.060e-03 | 7.373e-02 | 2.486e-01 | 1.660e-04 |
|---|
Expression data for subnetwork 1286 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
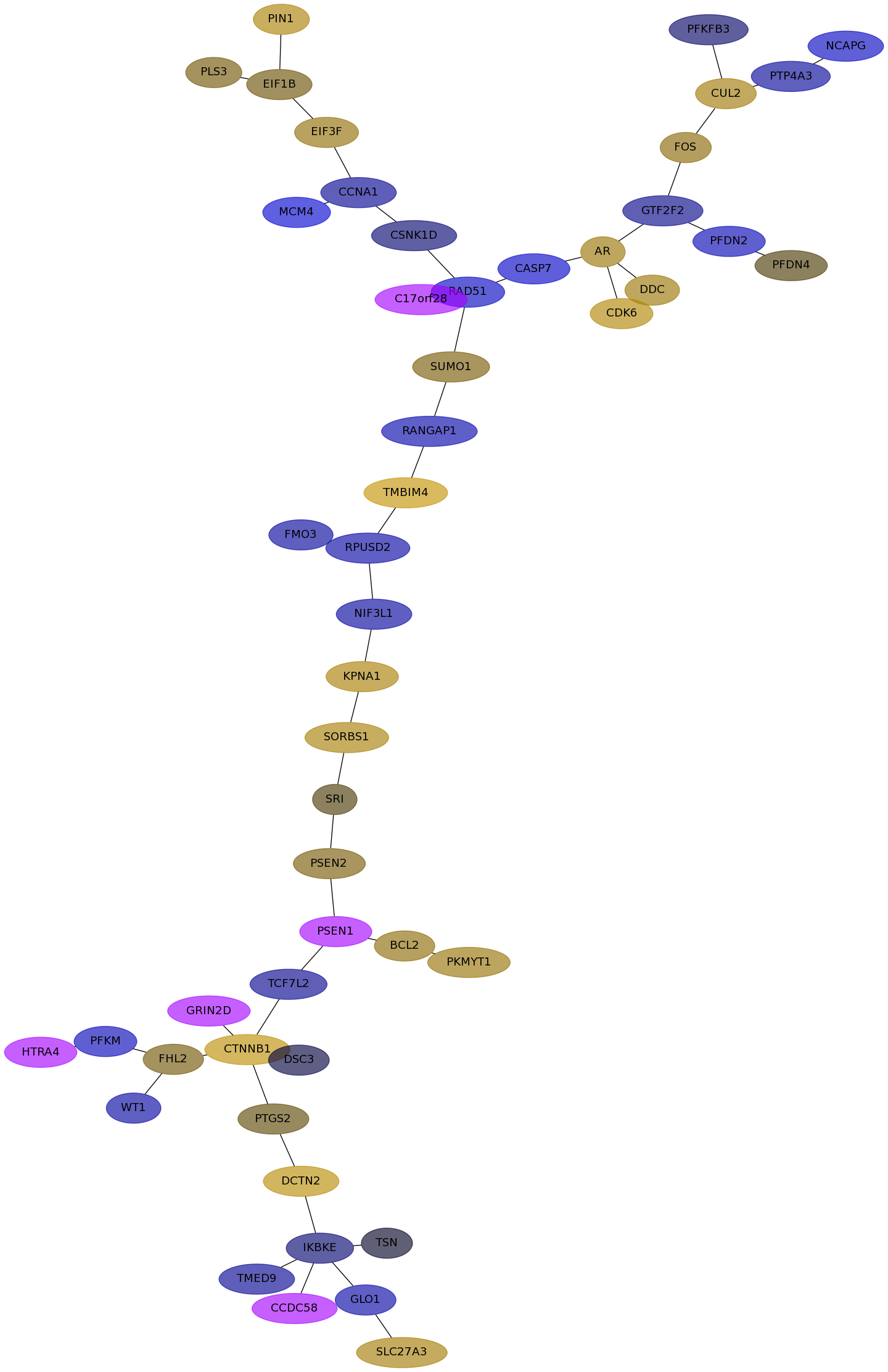
Score for each gene in subnetwork 1286 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| TMBIM4 |   | 1 | 652 | 1198 | 1207 | 0.251 | 0.197 | 0.017 | -0.160 | 0.169 |
|---|
| CCDC58 |   | 2 | 391 | 1198 | 1184 | undef | 0.241 | -0.030 | undef | undef |
|---|
| C17orf28 |   | 1 | 652 | 1198 | 1207 | undef | -0.094 | 0.094 | undef | undef |
|---|
| DCTN2 |   | 3 | 265 | 859 | 843 | 0.192 | 0.014 | 0.150 | 0.061 | 0.303 |
|---|
| PTP4A3 |   | 3 | 265 | 364 | 362 | -0.090 | 0.137 | -0.196 | 0.085 | -0.004 |
|---|
| RPUSD2 |   | 2 | 391 | 1198 | 1184 | -0.123 | 0.209 | -0.015 | 0.199 | 0.169 |
|---|
| NCAPG |   | 1 | 652 | 1198 | 1207 | -0.237 | 0.253 | -0.183 | 0.065 | 0.020 |
|---|
| PSEN1 |   | 5 | 148 | 847 | 821 | undef | -0.041 | 0.110 | undef | undef |
|---|
| EIF1B |   | 6 | 120 | 57 | 62 | 0.029 | 0.149 | 0.075 | 0.332 | -0.070 |
|---|
| SUMO1 |   | 3 | 265 | 1106 | 1083 | 0.038 | 0.245 | -0.007 | 0.144 | 0.118 |
|---|
| PLS3 |   | 2 | 391 | 57 | 80 | 0.034 | 0.144 | 0.175 | 0.247 | 0.256 |
|---|
| MCM4 |   | 1 | 652 | 1198 | 1207 | -0.344 | 0.141 | -0.085 | 0.037 | 0.030 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| GTF2F2 |   | 3 | 265 | 283 | 292 | -0.060 | -0.059 | -0.085 | 0.119 | -0.054 |
|---|
| SRI |   | 5 | 148 | 563 | 553 | 0.013 | 0.270 | -0.040 | 0.181 | 0.057 |
|---|
| PSEN2 |   | 2 | 391 | 1198 | 1184 | 0.039 | 0.239 | -0.068 | -0.157 | 0.045 |
|---|
| WT1 |   | 8 | 84 | 283 | 278 | -0.106 | 0.254 | -0.039 | 0.036 | 0.172 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| TMED9 |   | 2 | 391 | 1198 | 1184 | -0.076 | 0.142 | 0.057 | -0.078 | 0.321 |
|---|
| EIF3F |   | 1 | 652 | 1198 | 1207 | 0.073 | -0.167 | 0.092 | 0.128 | 0.035 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| FMO3 |   | 1 | 652 | 1198 | 1207 | -0.095 | -0.004 | -0.170 | 0.145 | -0.154 |
|---|
| DDC |   | 4 | 198 | 715 | 700 | 0.093 | -0.121 | 0.157 | -0.078 | 0.114 |
|---|
| NIF3L1 |   | 1 | 652 | 1198 | 1207 | -0.098 | 0.153 | -0.130 | -0.113 | 0.013 |
|---|
| DSC3 |   | 2 | 391 | 1198 | 1184 | -0.010 | -0.023 | 0.014 | 0.324 | 0.147 |
|---|
| TSN |   | 1 | 652 | 1198 | 1207 | -0.005 | 0.044 | -0.094 | 0.228 | 0.145 |
|---|
| FHL2 |   | 5 | 148 | 206 | 202 | 0.033 | 0.143 | 0.239 | -0.033 | 0.294 |
|---|
| PFKFB3 |   | 1 | 652 | 1198 | 1207 | -0.026 | 0.301 | 0.085 | 0.328 | 0.331 |
|---|
| RANGAP1 |   | 1 | 652 | 1198 | 1207 | -0.137 | 0.060 | 0.019 | 0.063 | 0.080 |
|---|
| GLO1 |   | 3 | 265 | 859 | 843 | -0.123 | 0.266 | -0.089 | 0.135 | 0.076 |
|---|
| PKMYT1 |   | 5 | 148 | 620 | 603 | 0.085 | 0.175 | -0.143 | 0.046 | -0.070 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| PFKM |   | 4 | 198 | 492 | 499 | -0.193 | -0.030 | -0.079 | 0.293 | 0.118 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| PTGS2 |   | 2 | 391 | 1198 | 1184 | 0.022 | 0.091 | -0.073 | 0.250 | 0.152 |
|---|
| KPNA1 |   | 3 | 265 | 1198 | 1173 | 0.130 | 0.007 | 0.152 | -0.083 | 0.086 |
|---|
| CASP7 |   | 8 | 84 | 283 | 278 | -0.270 | 0.216 | 0.095 | 0.255 | 0.035 |
|---|
| IKBKE |   | 8 | 84 | 756 | 726 | -0.033 | 0.227 | -0.041 | -0.095 | 0.079 |
|---|
| RAD51 |   | 4 | 198 | 696 | 684 | -0.225 | 0.146 | -0.098 | 0.135 | 0.061 |
|---|
| PFDN4 |   | 1 | 652 | 1198 | 1207 | 0.013 | 0.215 | -0.154 | 0.135 | 0.189 |
|---|
| CUL2 |   | 3 | 265 | 206 | 208 | 0.103 | 0.124 | -0.192 | -0.090 | 0.166 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| HTRA4 |   | 3 | 265 | 492 | 503 | undef | 0.102 | undef | undef | undef |
|---|
| AR |   | 9 | 77 | 283 | 277 | 0.089 | -0.163 | 0.081 | -0.035 | -0.030 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| PFDN2 |   | 1 | 652 | 1198 | 1207 | -0.167 | 0.251 | -0.171 | -0.049 | 0.126 |
|---|
| SLC27A3 |   | 2 | 391 | 756 | 756 | 0.117 | -0.019 | -0.027 | 0.058 | 0.028 |
|---|
| CSNK1D |   | 2 | 391 | 1198 | 1184 | -0.034 | -0.222 | -0.044 | 0.170 | 0.013 |
|---|
GO Enrichment output for subnetwork 1286 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 9.241E-08 | 2.125E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 3.113E-07 | 3.58E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.477E-06 | 4.199E-03 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 7.268E-06 | 4.179E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 1.597E-05 | 7.348E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 1.659E-05 | 6.358E-03 |
|---|
| negative regulation of binding | GO:0051100 |  | 2.362E-05 | 7.762E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 3.488E-05 | 0.01002819 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 3.488E-05 | 8.914E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 4.907E-05 | 0.01128568 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.33E-05 | 0.01114458 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.164E-10 | 5.287E-07 |
|---|
| protein sumoylation | GO:0016925 |  | 2.915E-08 | 3.561E-05 |
|---|
| Notch receptor processing | GO:0007220 |  | 4.123E-07 | 3.357E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.157E-07 | 2.539E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 8.225E-07 | 4.019E-04 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 1.436E-06 | 5.846E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 2.291E-06 | 7.996E-04 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 3.428E-06 | 1.047E-03 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 3.604E-06 | 9.783E-04 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 6.699E-06 | 1.637E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 7.622E-06 | 1.693E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.765E-10 | 4.248E-07 |
|---|
| protein sumoylation | GO:0016925 |  | 4.018E-08 | 4.833E-05 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.461E-07 | 3.578E-04 |
|---|
| Notch receptor processing | GO:0007220 |  | 5.253E-07 | 3.16E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.048E-06 | 5.041E-04 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 1.828E-06 | 7.332E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 2.917E-06 | 1.003E-03 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 4.364E-06 | 1.312E-03 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 5.661E-06 | 1.513E-03 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 8.525E-06 | 2.051E-03 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.044E-05 | 2.285E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 9.241E-08 | 2.125E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 3.113E-07 | 3.58E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.477E-06 | 4.199E-03 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 7.268E-06 | 4.179E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 1.597E-05 | 7.348E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 1.659E-05 | 6.358E-03 |
|---|
| negative regulation of binding | GO:0051100 |  | 2.362E-05 | 7.762E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 3.488E-05 | 0.01002819 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 3.488E-05 | 8.914E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 4.907E-05 | 0.01128568 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.33E-05 | 0.01114458 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 9.241E-08 | 2.125E-04 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 3.113E-07 | 3.58E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.477E-06 | 4.199E-03 |
|---|
| negative regulation of transcription regulator activity | GO:0090048 |  | 7.268E-06 | 4.179E-03 |
|---|
| focal adhesion formation | GO:0048041 |  | 1.597E-05 | 7.348E-03 |
|---|
| negative regulation of DNA binding | GO:0043392 |  | 1.659E-05 | 6.358E-03 |
|---|
| negative regulation of binding | GO:0051100 |  | 2.362E-05 | 7.762E-03 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 3.488E-05 | 0.01002819 |
|---|
| catecholamine biosynthetic process | GO:0042423 |  | 3.488E-05 | 8.914E-03 |
|---|
| regulation of protein localization | GO:0032880 |  | 4.907E-05 | 0.01128568 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.33E-05 | 0.01114458 |
|---|


