Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1266
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3371 | 1.268e-03 | 2.000e-05 | 1.506e-01 | 3.819e-09 |
|---|
| IPC | 0.3219 | 3.798e-01 | 2.064e-01 | 9.420e-01 | 7.385e-02 |
|---|
| Loi | 0.4877 | 3.200e-05 | 0.000e+00 | 4.793e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5962 | 1.000e-06 | 0.000e+00 | 1.207e-01 | 0.000e+00 |
|---|
| Wang | 0.2537 | 8.823e-03 | 7.253e-02 | 2.456e-01 | 1.571e-04 |
|---|
Expression data for subnetwork 1266 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
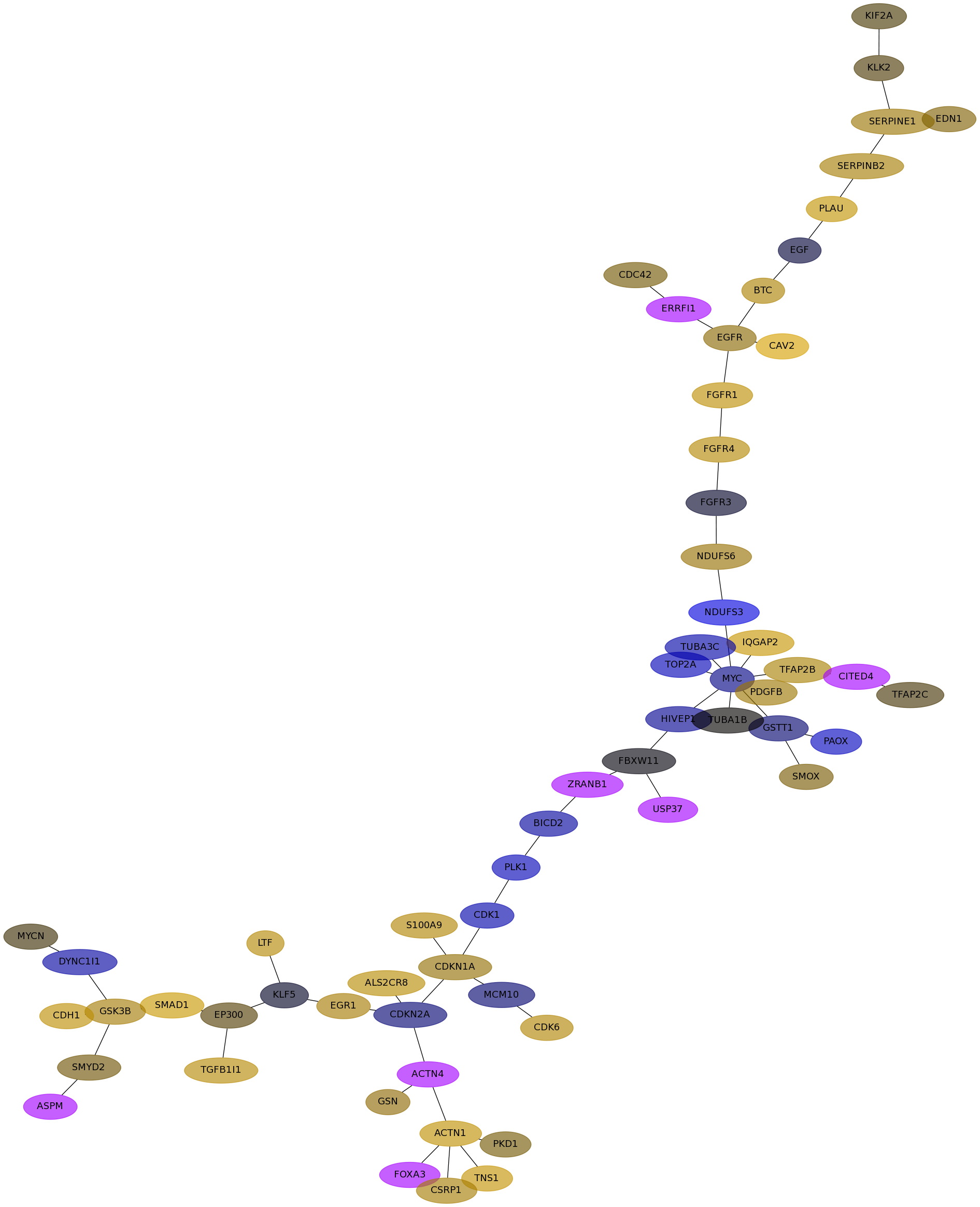
Score for each gene in subnetwork 1266 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| FBXW11 |   | 1 | 652 | 1144 | 1144 | -0.001 | 0.154 | 0.225 | 0.262 | 0.169 |
|---|
| BTC |   | 3 | 265 | 398 | 404 | 0.139 | 0.226 | 0.025 | -0.097 | -0.059 |
|---|
| MCM10 |   | 3 | 265 | 444 | 447 | -0.033 | 0.178 | -0.245 | 0.015 | -0.063 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| USP37 |   | 1 | 652 | 1144 | 1144 | undef | 0.075 | -0.159 | undef | undef |
|---|
| NDUFS6 |   | 1 | 652 | 1144 | 1144 | 0.084 | 0.261 | 0.068 | -0.021 | -0.038 |
|---|
| EGF |   | 11 | 59 | 398 | 383 | -0.009 | -0.212 | 0.182 | -0.276 | 0.005 |
|---|
| FOXA3 |   | 6 | 120 | 657 | 631 | undef | 0.192 | 0.289 | undef | undef |
|---|
| GSTT1 |   | 6 | 120 | 364 | 355 | -0.034 | 0.148 | 0.107 | -0.190 | -0.064 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| DYNC1I1 |   | 5 | 148 | 182 | 181 | -0.109 | -0.069 | 0.013 | -0.086 | 0.088 |
|---|
| PDGFB |   | 2 | 391 | 444 | 458 | 0.100 | 0.020 | 0.046 | 0.085 | 0.338 |
|---|
| LTF |   | 7 | 100 | 492 | 495 | 0.168 | -0.082 | 0.060 | -0.268 | -0.280 |
|---|
| SERPINB2 |   | 1 | 652 | 1144 | 1144 | 0.120 | 0.152 | -0.030 | 0.034 | -0.096 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| KLK2 |   | 1 | 652 | 1144 | 1144 | 0.014 | 0.204 | 0.002 | -0.115 | -0.048 |
|---|
| TOP2A |   | 2 | 391 | 444 | 458 | -0.185 | 0.193 | -0.168 | 0.054 | 0.071 |
|---|
| FGFR3 |   | 5 | 148 | 963 | 938 | -0.006 | 0.160 | 0.071 | 0.018 | -0.111 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| PLAU |   | 6 | 120 | 444 | 438 | 0.238 | 0.187 | 0.153 | 0.141 | 0.192 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| SMOX |   | 2 | 391 | 444 | 458 | 0.040 | -0.232 | 0.276 | -0.089 | 0.023 |
|---|
| MYCN |   | 3 | 265 | 859 | 843 | 0.009 | 0.125 | 0.057 | -0.003 | -0.084 |
|---|
| ALS2CR8 |   | 1 | 652 | 1144 | 1144 | 0.186 | -0.003 | 0.065 | 0.140 | -0.092 |
|---|
| ZRANB1 |   | 1 | 652 | 1144 | 1144 | undef | 0.205 | undef | undef | undef |
|---|
| ERRFI1 |   | 2 | 391 | 937 | 928 | undef | 0.076 | 0.215 | undef | undef |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| PLK1 |   | 5 | 148 | 444 | 440 | -0.179 | 0.142 | -0.143 | 0.094 | -0.163 |
|---|
| CSRP1 |   | 1 | 652 | 1144 | 1144 | 0.121 | -0.053 | 0.224 | 0.088 | 0.080 |
|---|
| CITED4 |   | 1 | 652 | 1144 | 1144 | undef | 0.172 | -0.054 | undef | undef |
|---|
| ASPM |   | 1 | 652 | 1144 | 1144 | undef | 0.175 | 0.019 | undef | undef |
|---|
| TFAP2B |   | 1 | 652 | 1144 | 1144 | 0.132 | -0.101 | 0.107 | -0.059 | -0.086 |
|---|
| EDN1 |   | 1 | 652 | 1144 | 1144 | 0.050 | 0.100 | 0.126 | 0.249 | 0.121 |
|---|
| TFAP2C |   | 1 | 652 | 1144 | 1144 | 0.012 | 0.026 | 0.076 | 0.025 | 0.044 |
|---|
| TNS1 |   | 1 | 652 | 1144 | 1144 | 0.251 | -0.125 | 0.149 | 0.120 | 0.210 |
|---|
| ACTN1 |   | 4 | 198 | 657 | 635 | 0.217 | 0.016 | 0.347 | 0.068 | 0.314 |
|---|
| BICD2 |   | 1 | 652 | 1144 | 1144 | -0.105 | -0.050 | 0.164 | 0.127 | 0.145 |
|---|
| SMYD2 |   | 4 | 198 | 148 | 162 | 0.032 | 0.260 | 0.068 | 0.179 | 0.191 |
|---|
| CDK1 |   | 16 | 34 | 206 | 194 | -0.138 | 0.265 | -0.195 | 0.106 | 0.031 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| TUBA1B |   | 7 | 100 | 182 | 180 | 0.001 | 0.116 | 0.094 | 0.249 | 0.128 |
|---|
| KIF2A |   | 1 | 652 | 1144 | 1144 | 0.014 | 0.173 | 0.001 | 0.052 | 0.023 |
|---|
| PAOX |   | 1 | 652 | 1144 | 1144 | -0.222 | 0.255 | -0.190 | 0.163 | -0.086 |
|---|
| GSN |   | 1 | 652 | 1144 | 1144 | 0.067 | 0.032 | 0.106 | 0.173 | 0.127 |
|---|
| TUBA3C |   | 1 | 652 | 1144 | 1144 | -0.132 | 0.207 | -0.014 | 0.118 | -0.040 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| ACTN4 |   | 6 | 120 | 859 | 834 | undef | 0.035 | 0.180 | undef | undef |
|---|
| NDUFS3 |   | 1 | 652 | 1144 | 1144 | -0.448 | 0.194 | -0.073 | 0.171 | -0.046 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| SERPINE1 |   | 1 | 652 | 1144 | 1144 | 0.092 | 0.160 | 0.200 | 0.056 | 0.030 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| CDKN2A |   | 10 | 67 | 364 | 351 | -0.037 | 0.263 | -0.113 | 0.111 | -0.105 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| HIVEP1 |   | 6 | 120 | 419 | 410 | -0.072 | -0.199 | 0.069 | 0.000 | -0.042 |
|---|
| PKD1 |   | 1 | 652 | 1144 | 1144 | 0.037 | -0.016 | 0.149 | -0.024 | -0.012 |
|---|
GO Enrichment output for subnetwork 1266 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of blood coagulation | GO:0030193 |  | 2.085E-09 | 4.796E-06 |
|---|
| regulation of coagulation | GO:0050818 |  | 6.317E-09 | 7.264E-06 |
|---|
| negative regulation of blood coagulation | GO:0030195 |  | 1.064E-08 | 8.157E-06 |
|---|
| negative regulation of coagulation | GO:0050819 |  | 3.244E-08 | 1.865E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.159E-07 | 9.93E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.572E-06 | 6.025E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.801E-06 | 5.918E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.801E-06 | 5.178E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.732E-06 | 6.982E-04 |
|---|
| cell growth | GO:0016049 |  | 2.732E-06 | 6.284E-04 |
|---|
| response to hypoxia | GO:0001666 |  | 3.467E-06 | 7.25E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of blood coagulation | GO:0030193 |  | 1.097E-09 | 2.679E-06 |
|---|
| regulation of coagulation | GO:0050818 |  | 2.79E-09 | 3.407E-06 |
|---|
| negative regulation of blood coagulation | GO:0030195 |  | 7.435E-09 | 6.055E-06 |
|---|
| negative regulation of coagulation | GO:0050819 |  | 1.844E-08 | 1.126E-05 |
|---|
| response to hypoxia | GO:0001666 |  | 5.544E-08 | 2.709E-05 |
|---|
| response to oxygen levels | GO:0070482 |  | 6.574E-08 | 2.677E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.37E-07 | 4.781E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 4.919E-07 | 1.502E-04 |
|---|
| stem cell division | GO:0017145 |  | 7.321E-07 | 1.987E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 1.124E-06 | 2.746E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.335E-06 | 2.964E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of blood coagulation | GO:0030193 |  | 1.252E-09 | 3.013E-06 |
|---|
| regulation of coagulation | GO:0050818 |  | 3.309E-09 | 3.981E-06 |
|---|
| negative regulation of blood coagulation | GO:0030195 |  | 1.033E-08 | 8.285E-06 |
|---|
| negative regulation of coagulation | GO:0050819 |  | 2.56E-08 | 1.54E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 8.616E-08 | 4.146E-05 |
|---|
| response to hypoxia | GO:0001666 |  | 8.628E-08 | 3.46E-05 |
|---|
| response to oxygen levels | GO:0070482 |  | 1.023E-07 | 3.515E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 6.04E-07 | 1.817E-04 |
|---|
| stem cell division | GO:0017145 |  | 8.948E-07 | 2.392E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 1.462E-06 | 3.519E-04 |
|---|
| negative regulation of epithelial cell proliferation | GO:0050680 |  | 1.463E-06 | 3.2E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of blood coagulation | GO:0030193 |  | 2.085E-09 | 4.796E-06 |
|---|
| regulation of coagulation | GO:0050818 |  | 6.317E-09 | 7.264E-06 |
|---|
| negative regulation of blood coagulation | GO:0030195 |  | 1.064E-08 | 8.157E-06 |
|---|
| negative regulation of coagulation | GO:0050819 |  | 3.244E-08 | 1.865E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.159E-07 | 9.93E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.572E-06 | 6.025E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.801E-06 | 5.918E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.801E-06 | 5.178E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.732E-06 | 6.982E-04 |
|---|
| cell growth | GO:0016049 |  | 2.732E-06 | 6.284E-04 |
|---|
| response to hypoxia | GO:0001666 |  | 3.467E-06 | 7.25E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of blood coagulation | GO:0030193 |  | 2.085E-09 | 4.796E-06 |
|---|
| regulation of coagulation | GO:0050818 |  | 6.317E-09 | 7.264E-06 |
|---|
| negative regulation of blood coagulation | GO:0030195 |  | 1.064E-08 | 8.157E-06 |
|---|
| negative regulation of coagulation | GO:0050819 |  | 3.244E-08 | 1.865E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.159E-07 | 9.93E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.572E-06 | 6.025E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.801E-06 | 5.918E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.801E-06 | 5.178E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 2.732E-06 | 6.982E-04 |
|---|
| cell growth | GO:0016049 |  | 2.732E-06 | 6.284E-04 |
|---|
| response to hypoxia | GO:0001666 |  | 3.467E-06 | 7.25E-04 |
|---|


