Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1247
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3348 | 1.409e-03 | 2.400e-05 | 1.577e-01 | 5.332e-09 |
|---|
| IPC | 0.3281 | 3.696e-01 | 1.971e-01 | 9.399e-01 | 6.846e-02 |
|---|
| Loi | 0.4865 | 3.400e-05 | 0.000e+00 | 4.849e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5986 | 1.000e-06 | 0.000e+00 | 1.156e-01 | 0.000e+00 |
|---|
| Wang | 0.2552 | 8.315e-03 | 6.989e-02 | 2.390e-01 | 1.389e-04 |
|---|
Expression data for subnetwork 1247 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
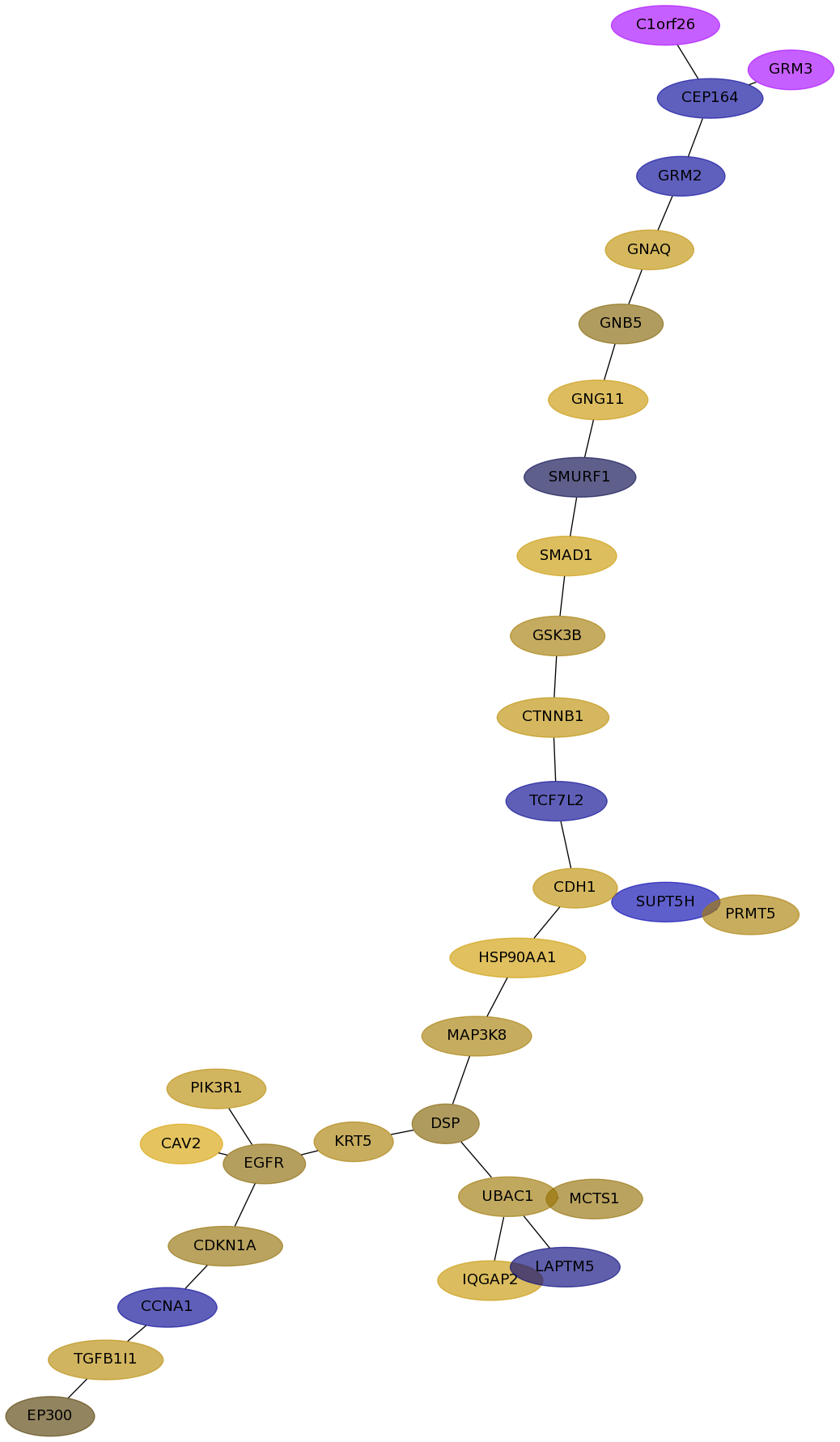
Score for each gene in subnetwork 1247 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| KRT5 |   | 5 | 148 | 923 | 898 | 0.129 | -0.125 | 0.046 | 0.064 | -0.190 |
|---|
| GNG11 |   | 4 | 198 | 33 | 41 | 0.270 | 0.049 | -0.039 | 0.135 | 0.106 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| MAP3K8 |   | 2 | 391 | 691 | 694 | 0.119 | 0.188 | -0.027 | 0.060 | -0.074 |
|---|
| UBAC1 |   | 2 | 391 | 598 | 600 | 0.101 | 0.003 | -0.088 | 0.008 | -0.040 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| SUPT5H |   | 9 | 77 | 1 | 18 | -0.150 | -0.085 | 0.007 | -0.298 | -0.068 |
|---|
| GRM3 |   | 1 | 652 | 930 | 943 | undef | -0.190 | 0.002 | 0.225 | -0.092 |
|---|
| DSP |   | 7 | 100 | 206 | 198 | 0.052 | 0.054 | 0.235 | -0.119 | 0.053 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| MCTS1 |   | 2 | 391 | 598 | 600 | 0.077 | 0.132 | 0.034 | 0.037 | 0.100 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| CEP164 |   | 1 | 652 | 930 | 943 | -0.090 | 0.076 | 0.301 | 0.210 | 0.256 |
|---|
| GNB5 |   | 1 | 652 | 930 | 943 | 0.053 | -0.059 | 0.026 | 0.064 | 0.103 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| LAPTM5 |   | 1 | 652 | 930 | 943 | -0.044 | 0.072 | -0.201 | -0.040 | -0.004 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| GNAQ |   | 2 | 391 | 412 | 423 | 0.221 | -0.090 | 0.226 | 0.105 | -0.062 |
|---|
| HSP90AA1 |   | 10 | 67 | 1 | 16 | 0.321 | -0.143 | 0.104 | 0.123 | -0.054 |
|---|
| PRMT5 |   | 1 | 652 | 930 | 943 | 0.127 | -0.058 | 0.016 | 0.144 | -0.132 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| SMURF1 |   | 9 | 77 | 33 | 34 | -0.013 | -0.222 | 0.236 | -0.049 | -0.006 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| C1orf26 |   | 1 | 652 | 930 | 943 | undef | -0.114 | 0.146 | undef | undef |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| GRM2 |   | 1 | 652 | 930 | 943 | -0.085 | -0.037 | 0.007 | 0.173 | -0.117 |
|---|
GO Enrichment output for subnetwork 1247 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.828E-09 | 1.111E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.255E-07 | 3.743E-04 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 5.875E-06 | 4.505E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.639E-05 | 9.424E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.639E-05 | 7.539E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.326E-05 | 8.915E-03 |
|---|
| regulation of adenylate cyclase activity | GO:0045761 |  | 8.022E-05 | 0.02635869 |
|---|
| regulation of cyclase activity | GO:0031279 |  | 8.437E-05 | 0.02425732 |
|---|
| forebrain neuron development | GO:0021884 |  | 8.628E-05 | 0.02205033 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 8.628E-05 | 0.0198453 |
|---|
| peptidyl-arginine modification | GO:0018195 |  | 8.628E-05 | 0.01804118 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.494E-14 | 2.319E-10 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.127E-07 | 2.599E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 3.019E-07 | 2.458E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 8.421E-07 | 5.143E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.277E-06 | 1.112E-03 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 2.468E-06 | 1.005E-03 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 3.284E-06 | 1.146E-03 |
|---|
| skeletal muscle tissue development | GO:0007519 |  | 5.484E-06 | 1.675E-03 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 6.756E-06 | 1.834E-03 |
|---|
| myoblast differentiation | GO:0045445 |  | 1.006E-05 | 2.458E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.205E-05 | 2.676E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.164E-14 | 1.243E-10 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.954E-07 | 2.351E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 3.324E-07 | 2.665E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 9.271E-07 | 5.577E-04 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.302E-06 | 1.108E-03 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 2.716E-06 | 1.089E-03 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 3.615E-06 | 1.243E-03 |
|---|
| skeletal muscle tissue development | GO:0007519 |  | 6.05E-06 | 1.82E-03 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 7.435E-06 | 1.988E-03 |
|---|
| myoblast differentiation | GO:0045445 |  | 1.107E-05 | 2.663E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.107E-05 | 2.421E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.828E-09 | 1.111E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.255E-07 | 3.743E-04 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 5.875E-06 | 4.505E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.639E-05 | 9.424E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.639E-05 | 7.539E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.326E-05 | 8.915E-03 |
|---|
| regulation of adenylate cyclase activity | GO:0045761 |  | 8.022E-05 | 0.02635869 |
|---|
| regulation of cyclase activity | GO:0031279 |  | 8.437E-05 | 0.02425732 |
|---|
| forebrain neuron development | GO:0021884 |  | 8.628E-05 | 0.02205033 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 8.628E-05 | 0.0198453 |
|---|
| peptidyl-arginine modification | GO:0018195 |  | 8.628E-05 | 0.01804118 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.828E-09 | 1.111E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.255E-07 | 3.743E-04 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 5.875E-06 | 4.505E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.639E-05 | 9.424E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.639E-05 | 7.539E-03 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 2.326E-05 | 8.915E-03 |
|---|
| regulation of adenylate cyclase activity | GO:0045761 |  | 8.022E-05 | 0.02635869 |
|---|
| regulation of cyclase activity | GO:0031279 |  | 8.437E-05 | 0.02425732 |
|---|
| forebrain neuron development | GO:0021884 |  | 8.628E-05 | 0.02205033 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 8.628E-05 | 0.0198453 |
|---|
| peptidyl-arginine modification | GO:0018195 |  | 8.628E-05 | 0.01804118 |
|---|


