Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1229
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3332 | 1.515e-03 | 2.700e-05 | 1.627e-01 | 6.655e-09 |
|---|
| IPC | 0.3373 | 3.546e-01 | 1.835e-01 | 9.368e-01 | 6.097e-02 |
|---|
| Loi | 0.4847 | 3.700e-05 | 0.000e+00 | 4.935e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6007 | 1.000e-06 | 0.000e+00 | 1.113e-01 | 0.000e+00 |
|---|
| Wang | 0.2566 | 7.873e-03 | 6.755e-02 | 2.331e-01 | 1.240e-04 |
|---|
Expression data for subnetwork 1229 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
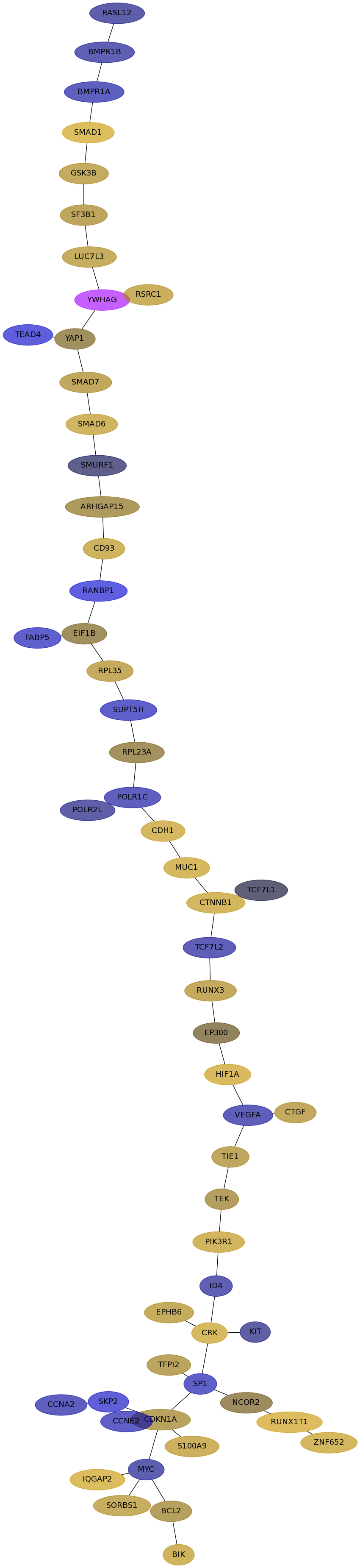
Score for each gene in subnetwork 1229 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CD93 |   | 1 | 652 | 665 | 687 | 0.169 | -0.102 | 0.036 | 0.160 | 0.254 |
|---|
| TEK |   | 7 | 100 | 148 | 155 | 0.063 | -0.149 | -0.049 | 0.174 | 0.113 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| CCNE2 |   | 8 | 84 | 536 | 515 | -0.123 | 0.136 | -0.178 | 0.241 | -0.055 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| BIK |   | 2 | 391 | 665 | 660 | 0.172 | -0.097 | 0.133 | -0.179 | -0.071 |
|---|
| SMAD7 |   | 3 | 265 | 665 | 651 | 0.099 | -0.035 | 0.131 | 0.169 | 0.147 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| EIF1B |   | 6 | 120 | 57 | 62 | 0.029 | 0.149 | 0.075 | 0.332 | -0.070 |
|---|
| RSRC1 |   | 2 | 391 | 606 | 609 | 0.140 | 0.023 | 0.104 | 0.105 | 0.212 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| CTGF |   | 10 | 67 | 57 | 52 | 0.106 | -0.169 | 0.181 | 0.126 | 0.209 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| RANBP1 |   | 1 | 652 | 665 | 687 | -0.326 | 0.063 | -0.066 | 0.199 | -0.048 |
|---|
| TEAD4 |   | 1 | 652 | 665 | 687 | -0.261 | 0.122 | -0.064 | 0.065 | 0.067 |
|---|
| BCL2 |   | 11 | 59 | 364 | 350 | 0.067 | -0.077 | -0.035 | 0.128 | 0.043 |
|---|
| EPHB6 |   | 7 | 100 | 419 | 407 | 0.122 | 0.008 | 0.042 | -0.219 | -0.059 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| SMAD6 |   | 4 | 198 | 665 | 650 | 0.173 | -0.241 | 0.053 | 0.266 | 0.070 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| RASL12 |   | 3 | 265 | 665 | 651 | -0.041 | -0.154 | 0.210 | 0.152 | 0.004 |
|---|
| TFPI2 |   | 1 | 652 | 665 | 687 | 0.080 | -0.218 | 0.078 | -0.089 | 0.047 |
|---|
| RPL35 |   | 5 | 148 | 57 | 63 | 0.114 | 0.108 | -0.012 | 0.370 | -0.220 |
|---|
| POLR2L |   | 3 | 265 | 57 | 71 | -0.038 | 0.037 | -0.012 | 0.224 | 0.015 |
|---|
| RUNX1T1 |   | 3 | 265 | 536 | 537 | 0.271 | -0.170 | -0.040 | 0.081 | -0.099 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| YAP1 |   | 2 | 391 | 665 | 660 | 0.030 | -0.199 | 0.132 | 0.262 | 0.075 |
|---|
| RUNX3 |   | 8 | 84 | 33 | 35 | 0.109 | 0.071 | -0.097 | 0.083 | -0.074 |
|---|
| FABP5 |   | 2 | 391 | 665 | 660 | -0.155 | 0.152 | -0.177 | 0.225 | 0.141 |
|---|
| SUPT5H |   | 9 | 77 | 1 | 18 | -0.150 | -0.085 | 0.007 | -0.298 | -0.068 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| ARHGAP15 |   | 1 | 652 | 665 | 687 | 0.050 | 0.004 | -0.235 | -0.022 | 0.013 |
|---|
| YWHAG |   | 2 | 391 | 606 | 609 | undef | 0.067 | 0.024 | undef | undef |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| TCF7L1 |   | 1 | 652 | 665 | 687 | -0.006 | -0.062 | -0.031 | 0.211 | -0.108 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| LUC7L3 |   | 13 | 47 | 148 | 148 | 0.123 | 0.043 | 0.237 | 0.175 | 0.074 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| ZNF652 |   | 3 | 265 | 536 | 537 | 0.208 | -0.215 | -0.045 | -0.150 | -0.076 |
|---|
| TIE1 |   | 6 | 120 | 148 | 156 | 0.094 | -0.108 | 0.005 | 0.141 | 0.252 |
|---|
| BMPR1A |   | 2 | 391 | 665 | 660 | -0.096 | -0.108 | 0.122 | 0.140 | -0.064 |
|---|
| POLR1C |   | 2 | 391 | 57 | 80 | -0.086 | 0.176 | -0.210 | 0.181 | 0.038 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| SMURF1 |   | 9 | 77 | 33 | 34 | -0.013 | -0.222 | 0.236 | -0.049 | -0.006 |
|---|
| BMPR1B |   | 1 | 652 | 665 | 687 | -0.059 | -0.037 | 0.078 | 0.163 | 0.146 |
|---|
GO Enrichment output for subnetwork 1229 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| BMP signaling pathway | GO:0030509 |  | 1.024E-08 | 2.356E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.932E-08 | 7.972E-05 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 1.446E-07 | 1.109E-04 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 5.561E-07 | 3.198E-04 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 6.227E-07 | 2.865E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 2.393E-06 | 9.172E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 3.587E-06 | 1.179E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.486E-06 | 1.29E-03 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 4.551E-06 | 1.163E-03 |
|---|
| regulation of ossification | GO:0030278 |  | 6.302E-06 | 1.449E-03 |
|---|
| cartilage development | GO:0051216 |  | 8.648E-06 | 1.808E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.733E-12 | 4.235E-09 |
|---|
| BMP signaling pathway | GO:0030509 |  | 3.172E-09 | 3.874E-06 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 3.658E-08 | 2.979E-05 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 1.169E-07 | 7.143E-05 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 1.213E-07 | 5.925E-05 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 1.3E-07 | 5.294E-05 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 3.312E-07 | 1.156E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 6.552E-07 | 2.001E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 8.362E-07 | 2.27E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.179E-06 | 2.881E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.264E-06 | 2.807E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.641E-12 | 3.949E-09 |
|---|
| BMP signaling pathway | GO:0030509 |  | 6.524E-09 | 7.849E-06 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 7.621E-08 | 6.112E-05 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 1.972E-07 | 1.186E-04 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 2.38E-07 | 1.145E-04 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 2.645E-07 | 1.061E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 5.38E-07 | 1.849E-04 |
|---|
| mesenchymal cell development | GO:0014031 |  | 1.048E-06 | 3.151E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 1.516E-06 | 4.053E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 1.823E-06 | 4.387E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.136E-06 | 4.671E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| BMP signaling pathway | GO:0030509 |  | 1.024E-08 | 2.356E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.932E-08 | 7.972E-05 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 1.446E-07 | 1.109E-04 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 5.561E-07 | 3.198E-04 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 6.227E-07 | 2.865E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 2.393E-06 | 9.172E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 3.587E-06 | 1.179E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.486E-06 | 1.29E-03 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 4.551E-06 | 1.163E-03 |
|---|
| regulation of ossification | GO:0030278 |  | 6.302E-06 | 1.449E-03 |
|---|
| cartilage development | GO:0051216 |  | 8.648E-06 | 1.808E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| BMP signaling pathway | GO:0030509 |  | 1.024E-08 | 2.356E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.932E-08 | 7.972E-05 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 1.446E-07 | 1.109E-04 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 5.561E-07 | 3.198E-04 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 6.227E-07 | 2.865E-04 |
|---|
| transforming growth factor beta receptor signaling pathway | GO:0007179 |  | 2.393E-06 | 9.172E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 3.587E-06 | 1.179E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.486E-06 | 1.29E-03 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 4.551E-06 | 1.163E-03 |
|---|
| regulation of ossification | GO:0030278 |  | 6.302E-06 | 1.449E-03 |
|---|
| cartilage development | GO:0051216 |  | 8.648E-06 | 1.808E-03 |
|---|


