Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1223
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3320 | 1.601e-03 | 2.900e-05 | 1.666e-01 | 7.734e-09 |
|---|
| IPC | 0.3395 | 3.509e-01 | 1.803e-01 | 9.361e-01 | 5.922e-02 |
|---|
| Loi | 0.4843 | 3.800e-05 | 0.000e+00 | 4.951e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6016 | 1.000e-06 | 0.000e+00 | 1.096e-01 | 0.000e+00 |
|---|
| Wang | 0.2571 | 7.705e-03 | 6.664e-02 | 2.308e-01 | 1.185e-04 |
|---|
Expression data for subnetwork 1223 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
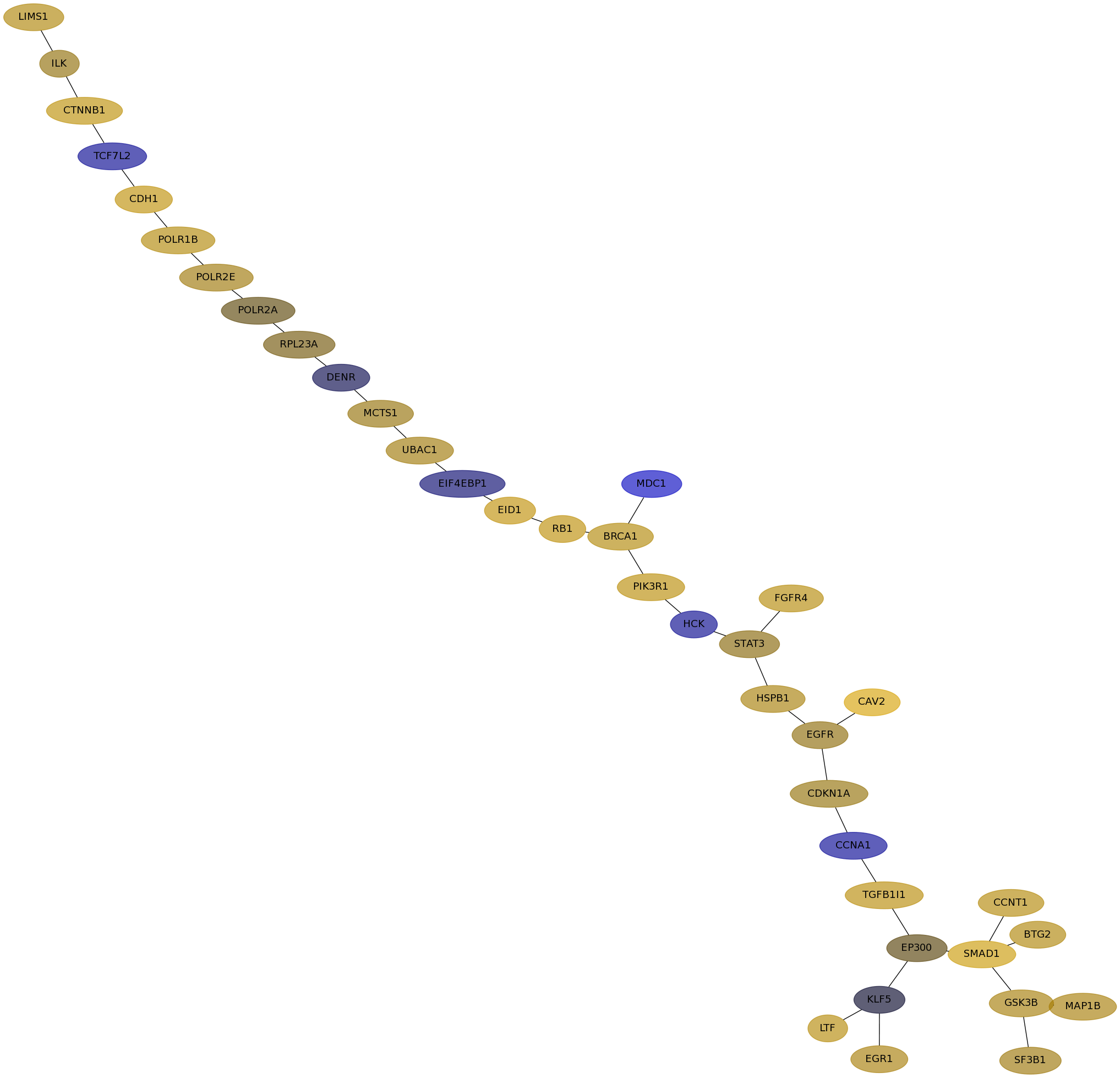
Score for each gene in subnetwork 1223 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| SF3B1 |   | 15 | 40 | 148 | 144 | 0.091 | -0.154 | 0.126 | -0.073 | -0.204 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| MDC1 |   | 2 | 391 | 598 | 600 | -0.213 | -0.001 | 0.067 | 0.138 | 0.179 |
|---|
| LTF |   | 7 | 100 | 492 | 495 | 0.168 | -0.082 | 0.060 | -0.268 | -0.280 |
|---|
| MCTS1 |   | 2 | 391 | 598 | 600 | 0.077 | 0.132 | 0.034 | 0.037 | 0.100 |
|---|
| RPL23A |   | 10 | 67 | 57 | 52 | 0.031 | 0.208 | -0.007 | 0.024 | 0.055 |
|---|
| DENR |   | 1 | 652 | 598 | 616 | -0.013 | 0.022 | -0.022 | -0.093 | 0.254 |
|---|
| RB1 |   | 26 | 18 | 271 | 263 | 0.200 | 0.013 | -0.051 | 0.057 | -0.082 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| STAT3 |   | 1 | 652 | 598 | 616 | 0.055 | -0.151 | -0.050 | -0.132 | -0.156 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| LIMS1 |   | 2 | 391 | 33 | 55 | 0.145 | 0.063 | 0.090 | -0.032 | 0.277 |
|---|
| EID1 |   | 4 | 198 | 552 | 547 | 0.212 | -0.069 | 0.051 | -0.010 | -0.037 |
|---|
| BTG2 |   | 6 | 120 | 1 | 21 | 0.146 | -0.029 | -0.093 | -0.111 | -0.170 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| UBAC1 |   | 2 | 391 | 598 | 600 | 0.101 | 0.003 | -0.088 | 0.008 | -0.040 |
|---|
| BRCA1 |   | 5 | 148 | 598 | 575 | 0.157 | -0.201 | -0.078 | -0.087 | 0.168 |
|---|
| ILK |   | 6 | 120 | 33 | 40 | 0.069 | -0.020 | 0.195 | 0.076 | 0.086 |
|---|
| HCK |   | 3 | 265 | 598 | 596 | -0.066 | -0.112 | -0.228 | -0.124 | -0.072 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| POLR2A |   | 5 | 148 | 271 | 267 | 0.019 | -0.107 | 0.247 | 0.091 | 0.141 |
|---|
| MAP1B |   | 13 | 47 | 148 | 148 | 0.118 | -0.123 | 0.263 | -0.107 | 0.100 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| EIF4EBP1 |   | 3 | 265 | 444 | 447 | -0.030 | 0.086 | -0.201 | -0.147 | 0.216 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| POLR2E |   | 4 | 198 | 598 | 577 | 0.095 | 0.127 | 0.039 | 0.166 | -0.018 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| SMAD1 |   | 24 | 21 | 33 | 30 | 0.287 | 0.049 | 0.159 | -0.062 | 0.048 |
|---|
| HSPB1 |   | 4 | 198 | 148 | 162 | 0.121 | 0.053 | 0.169 | 0.017 | 0.161 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
GO Enrichment output for subnetwork 1223 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.131E-08 | 2.601E-05 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.23E-07 | 1.414E-04 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 2.213E-06 | 1.697E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 2.298E-06 | 1.322E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 3.347E-06 | 1.54E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 4.167E-06 | 1.597E-03 |
|---|
| detection of bacterium | GO:0016045 |  | 5.514E-06 | 1.812E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 1.645E-05 | 4.729E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.811E-05 | 4.627E-03 |
|---|
| regulation of cytoskeleton organization | GO:0051493 |  | 2.275E-05 | 5.232E-03 |
|---|
| detection of biotic stimulus | GO:0009595 |  | 2.533E-05 | 5.296E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.718E-13 | 4.197E-10 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 8.16E-10 | 9.967E-07 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 2.99E-08 | 2.435E-05 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 4.661E-08 | 2.847E-05 |
|---|
| cell aging | GO:0007569 |  | 3.825E-07 | 1.869E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 4.04E-07 | 1.645E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 1.127E-06 | 3.932E-04 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.614E-06 | 4.93E-04 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 3.3E-06 | 8.958E-04 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 3.346E-06 | 8.174E-04 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 4.391E-06 | 9.753E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.338E-13 | 3.22E-10 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.165E-09 | 1.402E-06 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 3.808E-08 | 3.054E-05 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 6.061E-08 | 3.646E-05 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 5.302E-07 | 2.551E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 1.478E-06 | 5.927E-04 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.748E-06 | 6.009E-04 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 3.801E-06 | 1.143E-03 |
|---|
| positive regulation of peptide secretion | GO:0002793 |  | 4.327E-06 | 1.157E-03 |
|---|
| regulation of hormone metabolic process | GO:0032350 |  | 5.756E-06 | 1.385E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.155E-05 | 2.526E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.131E-08 | 2.601E-05 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.23E-07 | 1.414E-04 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 2.213E-06 | 1.697E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 2.298E-06 | 1.322E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 3.347E-06 | 1.54E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 4.167E-06 | 1.597E-03 |
|---|
| detection of bacterium | GO:0016045 |  | 5.514E-06 | 1.812E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 1.645E-05 | 4.729E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.811E-05 | 4.627E-03 |
|---|
| regulation of cytoskeleton organization | GO:0051493 |  | 2.275E-05 | 5.232E-03 |
|---|
| detection of biotic stimulus | GO:0009595 |  | 2.533E-05 | 5.296E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.131E-08 | 2.601E-05 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 1.23E-07 | 1.414E-04 |
|---|
| steroid hormone receptor signaling pathway | GO:0030518 |  | 2.213E-06 | 1.697E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 2.298E-06 | 1.322E-03 |
|---|
| intracellular receptor-mediated signaling pathway | GO:0030522 |  | 3.347E-06 | 1.54E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 4.167E-06 | 1.597E-03 |
|---|
| detection of bacterium | GO:0016045 |  | 5.514E-06 | 1.812E-03 |
|---|
| Wnt receptor signaling pathway | GO:0016055 |  | 1.645E-05 | 4.729E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.811E-05 | 4.627E-03 |
|---|
| regulation of cytoskeleton organization | GO:0051493 |  | 2.275E-05 | 5.232E-03 |
|---|
| detection of biotic stimulus | GO:0009595 |  | 2.533E-05 | 5.296E-03 |
|---|


