Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1221
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3320 | 1.602e-03 | 2.900e-05 | 1.666e-01 | 7.742e-09 |
|---|
| IPC | 0.3401 | 3.500e-01 | 1.795e-01 | 9.359e-01 | 5.881e-02 |
|---|
| Loi | 0.4841 | 3.800e-05 | 0.000e+00 | 4.962e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6018 | 1.000e-06 | 0.000e+00 | 1.091e-01 | 0.000e+00 |
|---|
| Wang | 0.2569 | 7.755e-03 | 6.692e-02 | 2.315e-01 | 1.201e-04 |
|---|
Expression data for subnetwork 1221 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
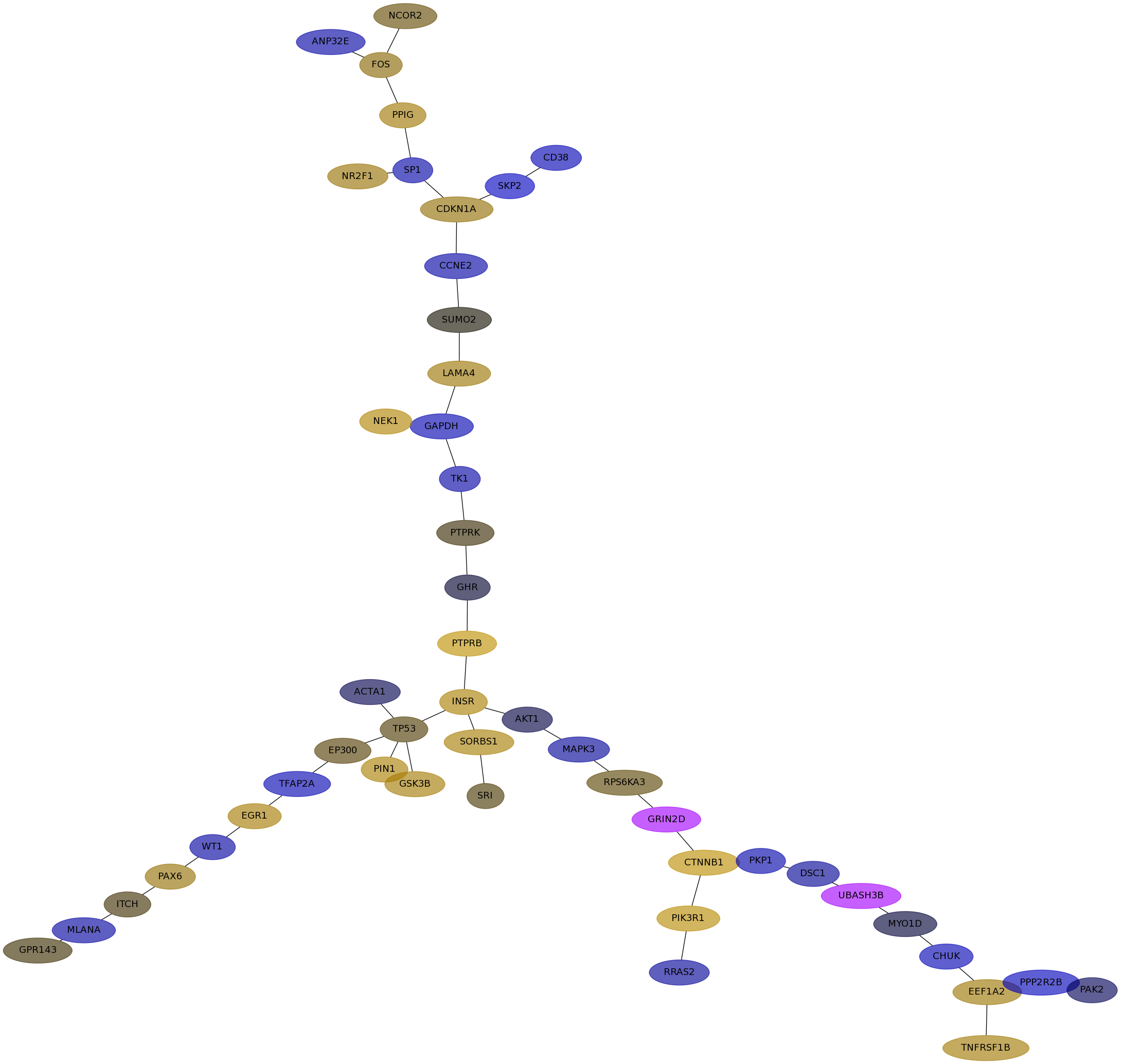
Score for each gene in subnetwork 1221 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CCNE2 |   | 8 | 84 | 536 | 515 | -0.123 | 0.136 | -0.178 | 0.241 | -0.055 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| MYO1D |   | 2 | 391 | 563 | 567 | -0.008 | 0.052 | 0.265 | -0.154 | 0.013 |
|---|
| NCOR2 |   | 11 | 59 | 283 | 274 | 0.024 | -0.096 | 0.211 | -0.163 | 0.114 |
|---|
| SORBS1 |   | 17 | 30 | 182 | 178 | 0.124 | -0.043 | 0.015 | 0.314 | 0.200 |
|---|
| PAX6 |   | 11 | 59 | 283 | 274 | 0.081 | 0.259 | -0.003 | 0.112 | 0.168 |
|---|
| EGR1 |   | 11 | 59 | 283 | 274 | 0.118 | -0.184 | 0.259 | 0.130 | 0.122 |
|---|
| UBASH3B |   | 1 | 652 | 563 | 584 | undef | 0.134 | 0.276 | undef | undef |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| GPR143 |   | 1 | 652 | 563 | 584 | 0.010 | 0.197 | -0.150 | 0.168 | -0.160 |
|---|
| CHUK |   | 1 | 652 | 563 | 584 | -0.172 | 0.173 | 0.080 | 0.152 | 0.215 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| PTPRB |   | 1 | 652 | 563 | 584 | 0.222 | -0.199 | 0.006 | 0.149 | 0.131 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| PPIG |   | 3 | 265 | 563 | 556 | 0.107 | -0.116 | 0.024 | 0.221 | 0.101 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| EEF1A2 |   | 9 | 77 | 182 | 179 | 0.098 | 0.032 | 0.220 | -0.119 | 0.102 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| GHR |   | 14 | 42 | 148 | 146 | -0.006 | -0.112 | 0.174 | 0.090 | 0.201 |
|---|
| TFAP2A |   | 2 | 391 | 563 | 567 | -0.162 | -0.070 | 0.146 | -0.095 | -0.009 |
|---|
| SRI |   | 5 | 148 | 563 | 553 | 0.013 | 0.270 | -0.040 | 0.181 | 0.057 |
|---|
| WT1 |   | 8 | 84 | 283 | 278 | -0.106 | 0.254 | -0.039 | 0.036 | 0.172 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| RRAS2 |   | 6 | 120 | 563 | 549 | -0.089 | 0.155 | 0.076 | 0.289 | 0.031 |
|---|
| SP1 |   | 16 | 34 | 283 | 272 | -0.129 | 0.222 | -0.254 | 0.239 | 0.082 |
|---|
| PTPRK |   | 6 | 120 | 148 | 156 | 0.008 | 0.156 | 0.141 | -0.094 | -0.024 |
|---|
| LAMA4 |   | 3 | 265 | 563 | 556 | 0.094 | -0.016 | 0.138 | 0.209 | 0.343 |
|---|
| TK1 |   | 1 | 652 | 563 | 584 | -0.114 | -0.089 | -0.156 | 0.029 | -0.052 |
|---|
| GAPDH |   | 2 | 391 | 563 | 567 | -0.153 | 0.159 | -0.040 | -0.053 | 0.029 |
|---|
| AKT1 |   | 13 | 47 | 444 | 430 | -0.012 | 0.045 | 0.126 | -0.099 | -0.168 |
|---|
| MLANA |   | 1 | 652 | 563 | 584 | -0.109 | 0.135 | -0.113 | 0.000 | 0.153 |
|---|
| ITCH |   | 1 | 652 | 563 | 584 | 0.010 | -0.097 | -0.150 | -0.170 | -0.120 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| CD38 |   | 1 | 652 | 563 | 584 | -0.173 | 0.186 | -0.256 | -0.189 | -0.142 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| NR2F1 |   | 6 | 120 | 444 | 438 | 0.086 | -0.252 | 0.113 | 0.133 | 0.207 |
|---|
| MAPK3 |   | 1 | 652 | 563 | 584 | -0.088 | 0.137 | -0.102 | -0.126 | -0.086 |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| NEK1 |   | 4 | 198 | 57 | 68 | 0.155 | 0.032 | 0.184 | 0.049 | 0.135 |
|---|
| ACTA1 |   | 1 | 652 | 563 | 584 | -0.015 | -0.090 | -0.041 | 0.145 | 0.132 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| ANP32E |   | 6 | 120 | 148 | 156 | -0.114 | 0.166 | -0.139 | 0.191 | 0.118 |
|---|
| PKP1 |   | 2 | 391 | 563 | 567 | -0.141 | 0.123 | 0.104 | 0.127 | 0.011 |
|---|
| DSC1 |   | 1 | 652 | 563 | 584 | -0.082 | -0.059 | -0.048 | -0.088 | -0.092 |
|---|
| RPS6KA3 |   | 1 | 652 | 563 | 584 | 0.021 | 0.268 | 0.132 | -0.179 | 0.255 |
|---|
GO Enrichment output for subnetwork 1221 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.132E-08 | 9.503E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.136E-07 | 1.306E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 7.146E-07 | 5.478E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.927E-06 | 1.108E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 2.385E-06 | 1.097E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.438E-06 | 9.346E-04 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 6.777E-06 | 2.227E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.777E-06 | 1.948E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 6.777E-06 | 1.732E-03 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 7.348E-06 | 1.69E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 9.818E-06 | 2.053E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.189E-08 | 5.347E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.763E-08 | 5.818E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.699E-08 | 4.641E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.867E-07 | 1.751E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 7.694E-07 | 3.759E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 8.531E-07 | 3.474E-04 |
|---|
| ER overload response | GO:0006983 |  | 2.703E-06 | 9.434E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 4.033E-06 | 1.232E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 5.761E-06 | 1.564E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 5.761E-06 | 1.407E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 7.9E-06 | 1.755E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.974E-08 | 7.155E-05 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.941E-08 | 5.944E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.68E-08 | 4.556E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 3.635E-07 | 2.186E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 9.648E-07 | 4.643E-04 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.111E-06 | 4.454E-04 |
|---|
| ER overload response | GO:0006983 |  | 3.405E-06 | 1.17E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 4.596E-06 | 1.382E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 7.253E-06 | 1.939E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 7.253E-06 | 1.745E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 9.944E-06 | 2.175E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.132E-08 | 9.503E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.136E-07 | 1.306E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 7.146E-07 | 5.478E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.927E-06 | 1.108E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 2.385E-06 | 1.097E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.438E-06 | 9.346E-04 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 6.777E-06 | 2.227E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.777E-06 | 1.948E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 6.777E-06 | 1.732E-03 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 7.348E-06 | 1.69E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 9.818E-06 | 2.053E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.132E-08 | 9.503E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.136E-07 | 1.306E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 7.146E-07 | 5.478E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.927E-06 | 1.108E-03 |
|---|
| response to insulin stimulus | GO:0032868 |  | 2.385E-06 | 1.097E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.438E-06 | 9.346E-04 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 6.777E-06 | 2.227E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.777E-06 | 1.948E-03 |
|---|
| regulation of embryonic development | GO:0045995 |  | 6.777E-06 | 1.732E-03 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 7.348E-06 | 1.69E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 9.818E-06 | 2.053E-03 |
|---|


