Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1214
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3327 | 1.554e-03 | 2.800e-05 | 1.645e-01 | 7.157e-09 |
|---|
| IPC | 0.3424 | 3.462e-01 | 1.762e-01 | 9.351e-01 | 5.704e-02 |
|---|
| Loi | 0.4831 | 4.000e-05 | 0.000e+00 | 5.010e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6013 | 1.000e-06 | 0.000e+00 | 1.101e-01 | 0.000e+00 |
|---|
| Wang | 0.2569 | 7.763e-03 | 6.696e-02 | 2.316e-01 | 1.204e-04 |
|---|
Expression data for subnetwork 1214 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
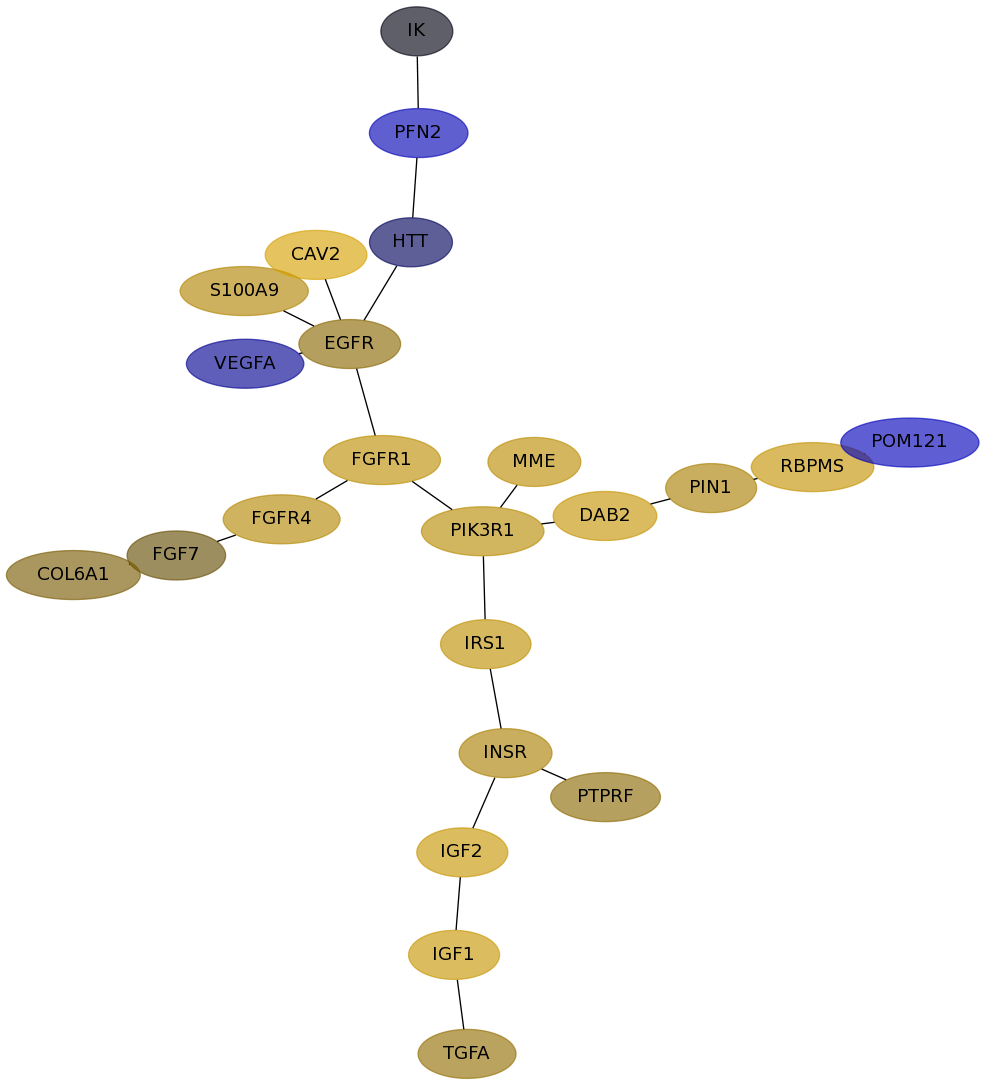
Score for each gene in subnetwork 1214 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| IK |   | 1 | 652 | 532 | 558 | -0.002 | 0.039 | 0.166 | 0.040 | -0.111 |
|---|
| TGFA |   | 8 | 84 | 439 | 429 | 0.077 | -0.057 | -0.027 | -0.158 | 0.020 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| PFN2 |   | 4 | 198 | 182 | 183 | -0.164 | 0.014 | -0.001 | 0.286 | -0.056 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| POM121 |   | 5 | 148 | 419 | 415 | -0.195 | -0.197 | 0.101 | 0.079 | 0.017 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| PTPRF |   | 2 | 391 | 439 | 452 | 0.066 | -0.111 | 0.190 | -0.132 | -0.251 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| HTT |   | 2 | 391 | 532 | 543 | -0.020 | -0.016 | -0.015 | 0.075 | -0.093 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| COL6A1 |   | 1 | 652 | 532 | 558 | 0.042 | -0.112 | 0.193 | -0.013 | 0.039 |
|---|
| IGF1 |   | 6 | 120 | 398 | 386 | 0.265 | -0.207 | -0.028 | 0.059 | -0.036 |
|---|
| FGF7 |   | 8 | 84 | 398 | 384 | 0.026 | -0.094 | 0.237 | 0.070 | 0.083 |
|---|
| RBPMS |   | 7 | 100 | 419 | 407 | 0.246 | 0.043 | 0.108 | 0.066 | -0.077 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| MME |   | 8 | 84 | 57 | 54 | 0.200 | -0.121 | 0.020 | 0.088 | -0.130 |
|---|
| IGF2 |   | 6 | 120 | 532 | 514 | 0.261 | -0.022 | 0.172 | 0.145 | 0.116 |
|---|
GO Enrichment output for subnetwork 1214 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 9.292E-09 | 2.137E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.767E-08 | 2.032E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.235E-08 | 1.714E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 3.976E-08 | 2.286E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.126E-07 | 5.182E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 3.804E-07 | 1.458E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.022E-07 | 1.322E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.999E-07 | 1.437E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.233E-07 | 1.593E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.353E-06 | 3.111E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.594E-06 | 3.333E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.828E-09 | 4.467E-06 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.479E-09 | 5.471E-06 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 4.602E-09 | 3.748E-06 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 6.565E-09 | 4.009E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 9.272E-09 | 4.53E-06 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.73E-08 | 1.112E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 8.209E-08 | 2.865E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.179E-07 | 3.602E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.528E-07 | 4.148E-05 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2.699E-07 | 6.593E-05 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 3.957E-07 | 8.788E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.207E-09 | 5.31E-06 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 4.601E-09 | 5.535E-06 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.41E-09 | 4.339E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.103E-08 | 6.634E-06 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 2.987E-08 | 1.437E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 9.234E-08 | 3.703E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.361E-07 | 4.68E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.494E-07 | 4.494E-05 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2.248E-07 | 6.009E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 3.866E-07 | 9.301E-05 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 3.896E-07 | 8.522E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 9.292E-09 | 2.137E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.767E-08 | 2.032E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.235E-08 | 1.714E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 3.976E-08 | 2.286E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.126E-07 | 5.182E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 3.804E-07 | 1.458E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.022E-07 | 1.322E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.999E-07 | 1.437E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.233E-07 | 1.593E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.353E-06 | 3.111E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.594E-06 | 3.333E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 9.292E-09 | 2.137E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.767E-08 | 2.032E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.235E-08 | 1.714E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 3.976E-08 | 2.286E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.126E-07 | 5.182E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 3.804E-07 | 1.458E-04 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.022E-07 | 1.322E-04 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.999E-07 | 1.437E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.233E-07 | 1.593E-04 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.353E-06 | 3.111E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.594E-06 | 3.333E-04 |
|---|


