Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1185
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3312 | 1.662e-03 | 3.100e-05 | 1.693e-01 | 8.722e-09 |
|---|
| IPC | 0.3454 | 3.413e-01 | 1.719e-01 | 9.341e-01 | 5.482e-02 |
|---|
| Loi | 0.4798 | 4.700e-05 | 0.000e+00 | 5.173e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6038 | 1.000e-06 | 0.000e+00 | 1.052e-01 | 0.000e+00 |
|---|
| Wang | 0.2571 | 7.700e-03 | 6.662e-02 | 2.307e-01 | 1.183e-04 |
|---|
Expression data for subnetwork 1185 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
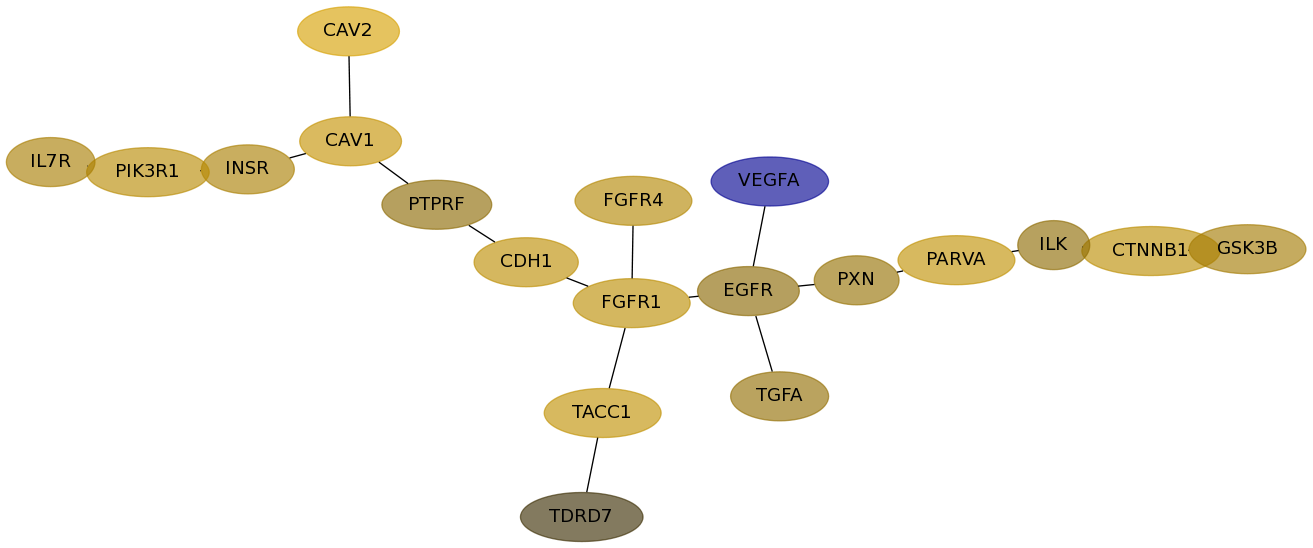
Score for each gene in subnetwork 1185 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| PXN |   | 5 | 148 | 439 | 432 | 0.085 | -0.088 | 0.284 | -0.022 | 0.114 |
|---|
| TACC1 |   | 5 | 148 | 326 | 330 | 0.229 | -0.051 | 0.020 | -0.022 | 0.126 |
|---|
| TGFA |   | 8 | 84 | 439 | 429 | 0.077 | -0.057 | -0.027 | -0.158 | 0.020 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PARVA |   | 1 | 652 | 439 | 473 | 0.224 | -0.214 | 0.082 | 0.114 | 0.263 |
|---|
| CAV1 |   | 10 | 67 | 439 | 428 | 0.251 | -0.092 | 0.124 | 0.079 | 0.119 |
|---|
| ILK |   | 6 | 120 | 33 | 40 | 0.069 | -0.020 | 0.195 | 0.076 | 0.086 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| PTPRF |   | 2 | 391 | 439 | 452 | 0.066 | -0.111 | 0.190 | -0.132 | -0.251 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| TDRD7 |   | 2 | 391 | 326 | 344 | 0.009 | 0.179 | 0.010 | -0.120 | -0.185 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 1185 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.518E-09 | 3.49E-06 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.699E-07 | 1.954E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.699E-07 | 1.302E-04 |
|---|
| vasculogenesis | GO:0001570 |  | 2.964E-07 | 1.704E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 3.391E-07 | 1.56E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 3.391E-07 | 1.3E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 3.391E-07 | 1.114E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 4.289E-07 | 1.233E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.924E-07 | 1.514E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 7.403E-07 | 1.703E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.46E-07 | 1.978E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.477E-10 | 8.494E-07 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 3.328E-09 | 4.065E-06 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 5.821E-08 | 4.741E-05 |
|---|
| vasculogenesis | GO:0001570 |  | 1.251E-07 | 7.643E-05 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.731E-07 | 8.459E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.032E-07 | 8.274E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 2.032E-07 | 7.092E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 3.247E-07 | 9.916E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 3.247E-07 | 8.815E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 3.247E-07 | 7.933E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 3.549E-07 | 7.882E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.367E-10 | 8.1E-07 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.577E-09 | 3.101E-06 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 6.759E-08 | 5.421E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.35E-07 | 8.12E-05 |
|---|
| vasculogenesis | GO:0001570 |  | 1.519E-07 | 7.312E-05 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 1.793E-07 | 7.191E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.359E-07 | 8.109E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.359E-07 | 7.096E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.359E-07 | 6.307E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 4.21E-07 | 1.013E-04 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 5.647E-07 | 1.235E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.518E-09 | 3.49E-06 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.699E-07 | 1.954E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.699E-07 | 1.302E-04 |
|---|
| vasculogenesis | GO:0001570 |  | 2.964E-07 | 1.704E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 3.391E-07 | 1.56E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 3.391E-07 | 1.3E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 3.391E-07 | 1.114E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 4.289E-07 | 1.233E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.924E-07 | 1.514E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 7.403E-07 | 1.703E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.46E-07 | 1.978E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.518E-09 | 3.49E-06 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 1.699E-07 | 1.954E-04 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.699E-07 | 1.302E-04 |
|---|
| vasculogenesis | GO:0001570 |  | 2.964E-07 | 1.704E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 3.391E-07 | 1.56E-04 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 3.391E-07 | 1.3E-04 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 3.391E-07 | 1.114E-04 |
|---|
| regulation of endothelial cell proliferation | GO:0001936 |  | 4.289E-07 | 1.233E-04 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 5.924E-07 | 1.514E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 7.403E-07 | 1.703E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 9.46E-07 | 1.978E-04 |
|---|


