Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1183
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3311 | 1.665e-03 | 3.100e-05 | 1.694e-01 | 8.745e-09 |
|---|
| IPC | 0.3460 | 3.403e-01 | 1.711e-01 | 9.339e-01 | 5.437e-02 |
|---|
| Loi | 0.4795 | 4.700e-05 | 0.000e+00 | 5.185e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6041 | 1.000e-06 | 0.000e+00 | 1.047e-01 | 0.000e+00 |
|---|
| Wang | 0.2572 | 7.665e-03 | 6.643e-02 | 2.302e-01 | 1.172e-04 |
|---|
Expression data for subnetwork 1183 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
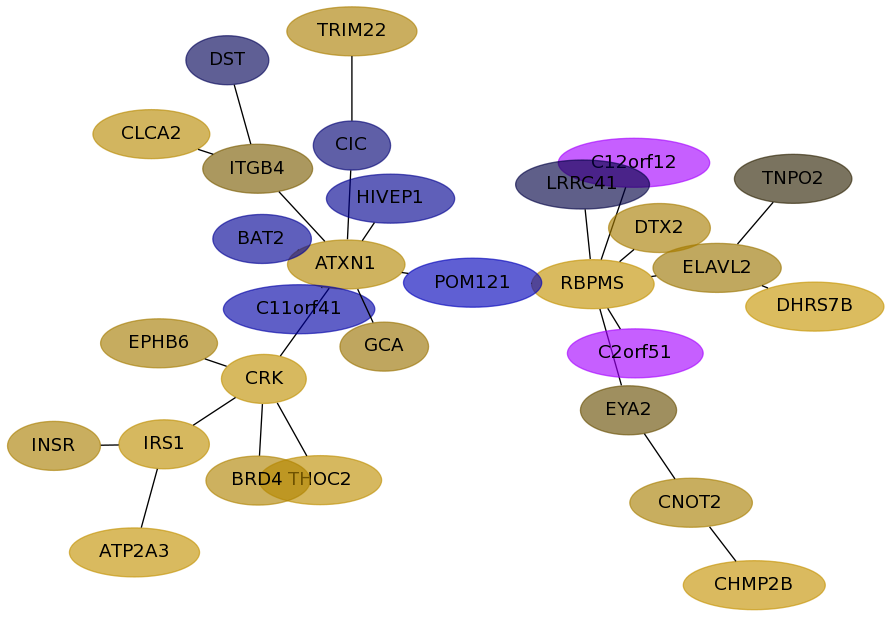
Score for each gene in subnetwork 1183 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| GCA |   | 2 | 391 | 419 | 433 | 0.092 | 0.022 | -0.143 | -0.202 | 0.180 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| C2orf51 |   | 1 | 652 | 419 | 453 | undef | -0.001 | 0.105 | undef | undef |
|---|
| BAT2 |   | 1 | 652 | 419 | 453 | -0.083 | -0.017 | 0.031 | 0.058 | 0.016 |
|---|
| CIC |   | 1 | 652 | 419 | 453 | -0.037 | -0.091 | 0.128 | -0.144 | -0.113 |
|---|
| C11orf41 |   | 2 | 391 | 364 | 375 | -0.126 | 0.062 | 0.058 | 0.045 | -0.157 |
|---|
| ATP2A3 |   | 9 | 77 | 1 | 18 | 0.240 | -0.040 | 0.082 | -0.229 | -0.025 |
|---|
| C12orf12 |   | 3 | 265 | 419 | 419 | undef | 0.135 | 0.002 | undef | undef |
|---|
| ATXN1 |   | 6 | 120 | 364 | 355 | 0.171 | 0.026 | 0.192 | 0.175 | 0.163 |
|---|
| TRIM22 |   | 1 | 652 | 419 | 453 | 0.138 | -0.091 | -0.102 | -0.114 | -0.113 |
|---|
| CHMP2B |   | 3 | 265 | 419 | 419 | 0.250 | 0.128 | 0.134 | -0.039 | 0.067 |
|---|
| POM121 |   | 5 | 148 | 419 | 415 | -0.195 | -0.197 | 0.101 | 0.079 | 0.017 |
|---|
| CNOT2 |   | 3 | 265 | 419 | 419 | 0.132 | 0.202 | 0.114 | 0.112 | 0.123 |
|---|
| IRS1 |   | 14 | 42 | 1 | 14 | 0.217 | -0.219 | 0.065 | 0.165 | 0.064 |
|---|
| DST |   | 2 | 391 | 419 | 433 | -0.019 | -0.094 | 0.343 | 0.065 | 0.194 |
|---|
| ELAVL2 |   | 4 | 198 | 419 | 417 | 0.096 | 0.085 | 0.115 | 0.158 | -0.076 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| CLCA2 |   | 5 | 148 | 206 | 202 | 0.188 | -0.176 | 0.243 | -0.153 | -0.000 |
|---|
| TNPO2 |   | 1 | 652 | 419 | 453 | 0.006 | -0.123 | 0.052 | 0.094 | 0.029 |
|---|
| DTX2 |   | 2 | 391 | 419 | 433 | 0.130 | 0.005 | 0.174 | -0.122 | -0.200 |
|---|
| EPHB6 |   | 7 | 100 | 419 | 407 | 0.122 | 0.008 | 0.042 | -0.219 | -0.059 |
|---|
| ITGB4 |   | 6 | 120 | 364 | 355 | 0.044 | 0.021 | 0.313 | -0.134 | -0.114 |
|---|
| RBPMS |   | 7 | 100 | 419 | 407 | 0.246 | 0.043 | 0.108 | 0.066 | -0.077 |
|---|
| EYA2 |   | 5 | 148 | 419 | 415 | 0.028 | -0.034 | 0.113 | -0.142 | -0.019 |
|---|
| HIVEP1 |   | 6 | 120 | 419 | 410 | -0.072 | -0.199 | 0.069 | 0.000 | -0.042 |
|---|
| LRRC41 |   | 3 | 265 | 419 | 419 | -0.012 | -0.136 | 0.210 | -0.080 | -0.125 |
|---|
| DHRS7B |   | 4 | 198 | 419 | 417 | 0.261 | 0.031 | -0.058 | -0.114 | 0.009 |
|---|
GO Enrichment output for subnetwork 1183 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.691E-05 | 0.13089436 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 9.66E-05 | 0.11108648 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 9.66E-05 | 0.07405765 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.446E-04 | 0.08314656 |
|---|
| male sex determination | GO:0030238 |  | 1.446E-04 | 0.06651724 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.646E-04 | 0.06307767 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.02E-04 | 0.0663829 |
|---|
| positive regulation of mesenchymal cell proliferation | GO:0002053 |  | 2.02E-04 | 0.05808504 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.02E-04 | 0.05163115 |
|---|
| regulation of mesenchymal cell proliferation | GO:0010464 |  | 2.688E-04 | 0.06183227 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.688E-04 | 0.05621115 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.449E-05 | 0.08425029 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.363E-05 | 0.06550649 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 8.032E-05 | 0.06540702 |
|---|
| cytoplasmic microtubule organization | GO:0031122 |  | 8.032E-05 | 0.04905526 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 9.764E-05 | 0.04770476 |
|---|
| male sex determination | GO:0030238 |  | 1.123E-04 | 0.0457154 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.495E-04 | 0.05216686 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.495E-04 | 0.045646 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 1.495E-04 | 0.04057422 |
|---|
| response to insulin stimulus | GO:0032868 |  | 1.848E-04 | 0.04515767 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 1.919E-04 | 0.04261719 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 3.555E-05 | 0.08552705 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 5.927E-05 | 0.07130718 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 8.877E-05 | 0.07119358 |
|---|
| male sex determination | GO:0030238 |  | 8.877E-05 | 0.05339519 |
|---|
| cytoplasmic microtubule organization | GO:0031122 |  | 8.877E-05 | 0.04271615 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.042E-04 | 0.04177985 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.241E-04 | 0.04264811 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 1.241E-04 | 0.0373171 |
|---|
| positive regulation of mesenchymal cell proliferation | GO:0002053 |  | 1.652E-04 | 0.04415724 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.652E-04 | 0.03974152 |
|---|
| response to insulin stimulus | GO:0032868 |  | 2.003E-04 | 0.04382105 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.691E-05 | 0.13089436 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 9.66E-05 | 0.11108648 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 9.66E-05 | 0.07405765 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.446E-04 | 0.08314656 |
|---|
| male sex determination | GO:0030238 |  | 1.446E-04 | 0.06651724 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.646E-04 | 0.06307767 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.02E-04 | 0.0663829 |
|---|
| positive regulation of mesenchymal cell proliferation | GO:0002053 |  | 2.02E-04 | 0.05808504 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.02E-04 | 0.05163115 |
|---|
| regulation of mesenchymal cell proliferation | GO:0010464 |  | 2.688E-04 | 0.06183227 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.688E-04 | 0.05621115 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 5.691E-05 | 0.13089436 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 9.66E-05 | 0.11108648 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 9.66E-05 | 0.07405765 |
|---|
| cell fate commitment involved in the formation of primary germ layers | GO:0060795 |  | 1.446E-04 | 0.08314656 |
|---|
| male sex determination | GO:0030238 |  | 1.446E-04 | 0.06651724 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 1.646E-04 | 0.06307767 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.02E-04 | 0.0663829 |
|---|
| positive regulation of mesenchymal cell proliferation | GO:0002053 |  | 2.02E-04 | 0.05808504 |
|---|
| protein heterooligomerization | GO:0051291 |  | 2.02E-04 | 0.05163115 |
|---|
| regulation of mesenchymal cell proliferation | GO:0010464 |  | 2.688E-04 | 0.06183227 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.688E-04 | 0.05621115 |
|---|


