Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1181
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3305 | 1.712e-03 | 3.200e-05 | 1.715e-01 | 9.394e-09 |
|---|
| IPC | 0.3476 | 3.376e-01 | 1.687e-01 | 9.333e-01 | 5.317e-02 |
|---|
| Loi | 0.4791 | 4.800e-05 | 0.000e+00 | 5.206e-02 | 0.000e+00 |
|---|
| Schmidt | 0.6049 | 1.000e-06 | 0.000e+00 | 1.032e-01 | 0.000e+00 |
|---|
| Wang | 0.2575 | 7.573e-03 | 6.593e-02 | 2.289e-01 | 1.143e-04 |
|---|
Expression data for subnetwork 1181 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
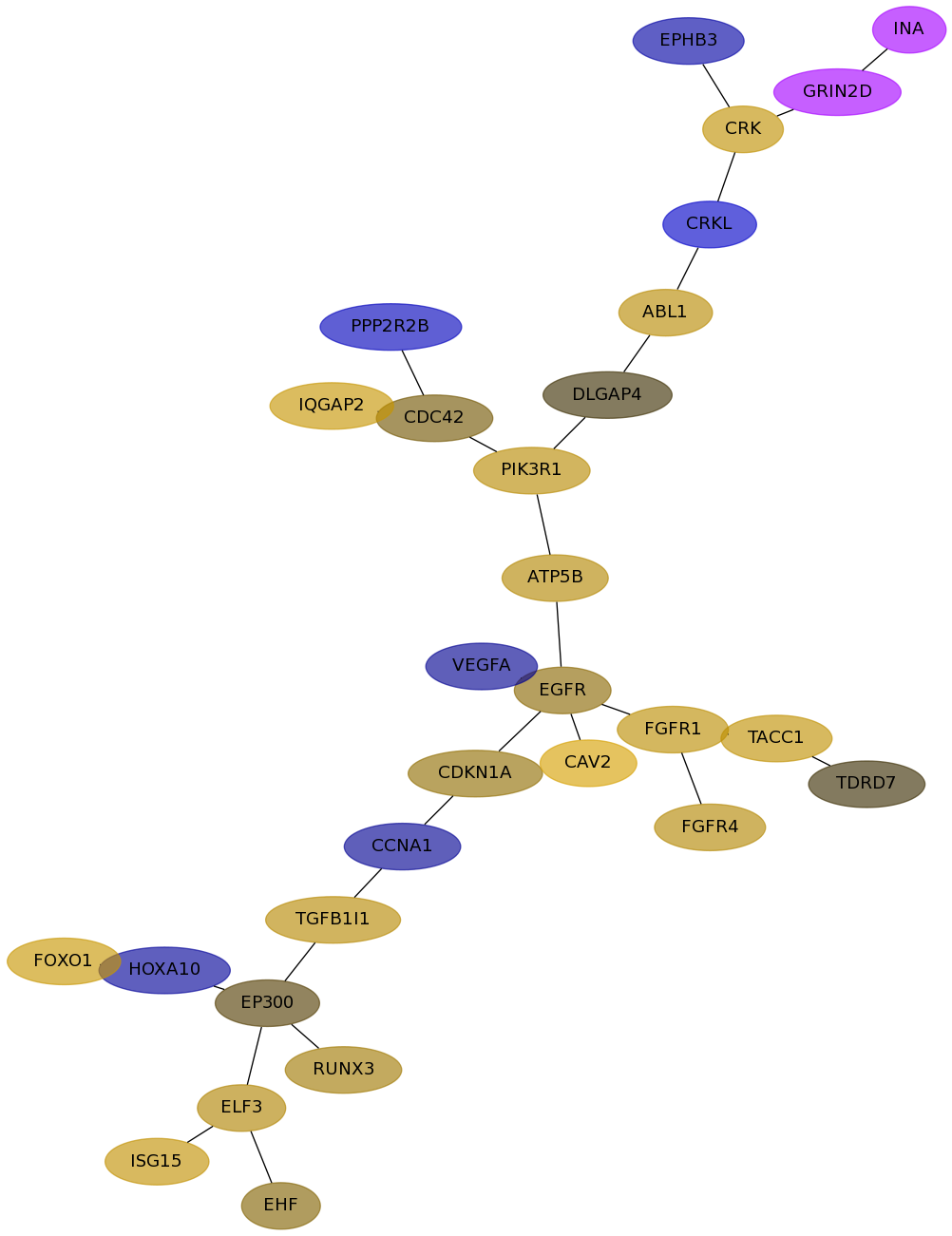
Score for each gene in subnetwork 1181 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| FOXO1 |   | 4 | 198 | 148 | 162 | 0.278 | 0.072 | 0.094 | -0.021 | 0.221 |
|---|
| TACC1 |   | 5 | 148 | 326 | 330 | 0.229 | -0.051 | 0.020 | -0.022 | 0.126 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| DLGAP4 |   | 5 | 148 | 326 | 330 | 0.010 | -0.029 | 0.180 | -0.201 | -0.043 |
|---|
| RUNX3 |   | 8 | 84 | 33 | 35 | 0.109 | 0.071 | -0.097 | 0.083 | -0.074 |
|---|
| ATP5B |   | 2 | 391 | 326 | 344 | 0.167 | -0.059 | -0.003 | 0.107 | 0.003 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| ISG15 |   | 2 | 391 | 326 | 344 | 0.235 | 0.227 | -0.032 | -0.160 | -0.133 |
|---|
| INA |   | 2 | 391 | 57 | 80 | undef | 0.144 | 0.025 | undef | undef |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| ELF3 |   | 2 | 391 | 326 | 344 | 0.154 | 0.178 | -0.022 | -0.052 | -0.076 |
|---|
| HOXA10 |   | 1 | 652 | 326 | 360 | -0.088 | 0.046 | 0.203 | -0.057 | 0.016 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| ABL1 |   | 8 | 84 | 326 | 316 | 0.193 | -0.152 | 0.200 | 0.108 | -0.059 |
|---|
| CRKL |   | 7 | 100 | 326 | 317 | -0.271 | -0.023 | 0.056 | 0.271 | -0.008 |
|---|
| EHF |   | 1 | 652 | 326 | 360 | 0.053 | -0.217 | -0.025 | 0.007 | -0.084 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| TDRD7 |   | 2 | 391 | 326 | 344 | 0.009 | 0.179 | 0.010 | -0.120 | -0.185 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| EPHB3 |   | 4 | 198 | 326 | 333 | -0.115 | -0.048 | 0.135 | 0.099 | -0.008 |
|---|
GO Enrichment output for subnetwork 1181 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 7.128E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 3.564E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 4.742E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 6.209E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.08E-06 | 4.967E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.58E-06 | 9.889E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.677E-06 | 1.208E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 5.045E-06 | 1.45E-03 |
|---|
| tissue remodeling | GO:0048771 |  | 5.153E-06 | 1.317E-03 |
|---|
| gland morphogenesis | GO:0022612 |  | 6.711E-06 | 1.543E-03 |
|---|
| microspike assembly | GO:0030035 |  | 6.711E-06 | 1.403E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 2.355E-07 | 5.753E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.355E-07 | 2.877E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.572E-07 | 5.352E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.572E-07 | 4.014E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 9.841E-07 | 4.809E-04 |
|---|
| tissue remodeling | GO:0048771 |  | 1.639E-06 | 6.675E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.927E-06 | 6.724E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 2.565E-06 | 7.832E-04 |
|---|
| gland morphogenesis | GO:0022612 |  | 3.329E-06 | 9.035E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 7.861E-06 | 1.921E-03 |
|---|
| microspike assembly | GO:0030035 |  | 9.418E-06 | 2.092E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 2.951E-07 | 7.099E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.951E-07 | 3.55E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.154E-07 | 4.134E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 5.154E-07 | 3.1E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 8.232E-07 | 3.961E-04 |
|---|
| tissue remodeling | GO:0048771 |  | 2.201E-06 | 8.825E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.413E-06 | 8.292E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.211E-06 | 9.657E-04 |
|---|
| gland morphogenesis | GO:0022612 |  | 3.211E-06 | 8.584E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 8.115E-06 | 1.952E-03 |
|---|
| microspike assembly | GO:0030035 |  | 9.836E-06 | 2.151E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 7.128E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 3.564E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 4.742E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 6.209E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.08E-06 | 4.967E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.58E-06 | 9.889E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.677E-06 | 1.208E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 5.045E-06 | 1.45E-03 |
|---|
| tissue remodeling | GO:0048771 |  | 5.153E-06 | 1.317E-03 |
|---|
| gland morphogenesis | GO:0022612 |  | 6.711E-06 | 1.543E-03 |
|---|
| microspike assembly | GO:0030035 |  | 6.711E-06 | 1.403E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 7.128E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 3.564E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 4.742E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 6.209E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.08E-06 | 4.967E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.58E-06 | 9.889E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.677E-06 | 1.208E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 5.045E-06 | 1.45E-03 |
|---|
| tissue remodeling | GO:0048771 |  | 5.153E-06 | 1.317E-03 |
|---|
| gland morphogenesis | GO:0022612 |  | 6.711E-06 | 1.543E-03 |
|---|
| microspike assembly | GO:0030035 |  | 6.711E-06 | 1.403E-03 |
|---|


