Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1158
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3325 | 1.564e-03 | 2.800e-05 | 1.649e-01 | 7.222e-09 |
|---|
| IPC | 0.3421 | 3.467e-01 | 1.766e-01 | 9.352e-01 | 5.725e-02 |
|---|
| Loi | 0.4773 | 5.300e-05 | 0.000e+00 | 5.294e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5998 | 1.000e-06 | 0.000e+00 | 1.132e-01 | 0.000e+00 |
|---|
| Wang | 0.2569 | 7.759e-03 | 6.694e-02 | 2.315e-01 | 1.202e-04 |
|---|
Expression data for subnetwork 1158 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
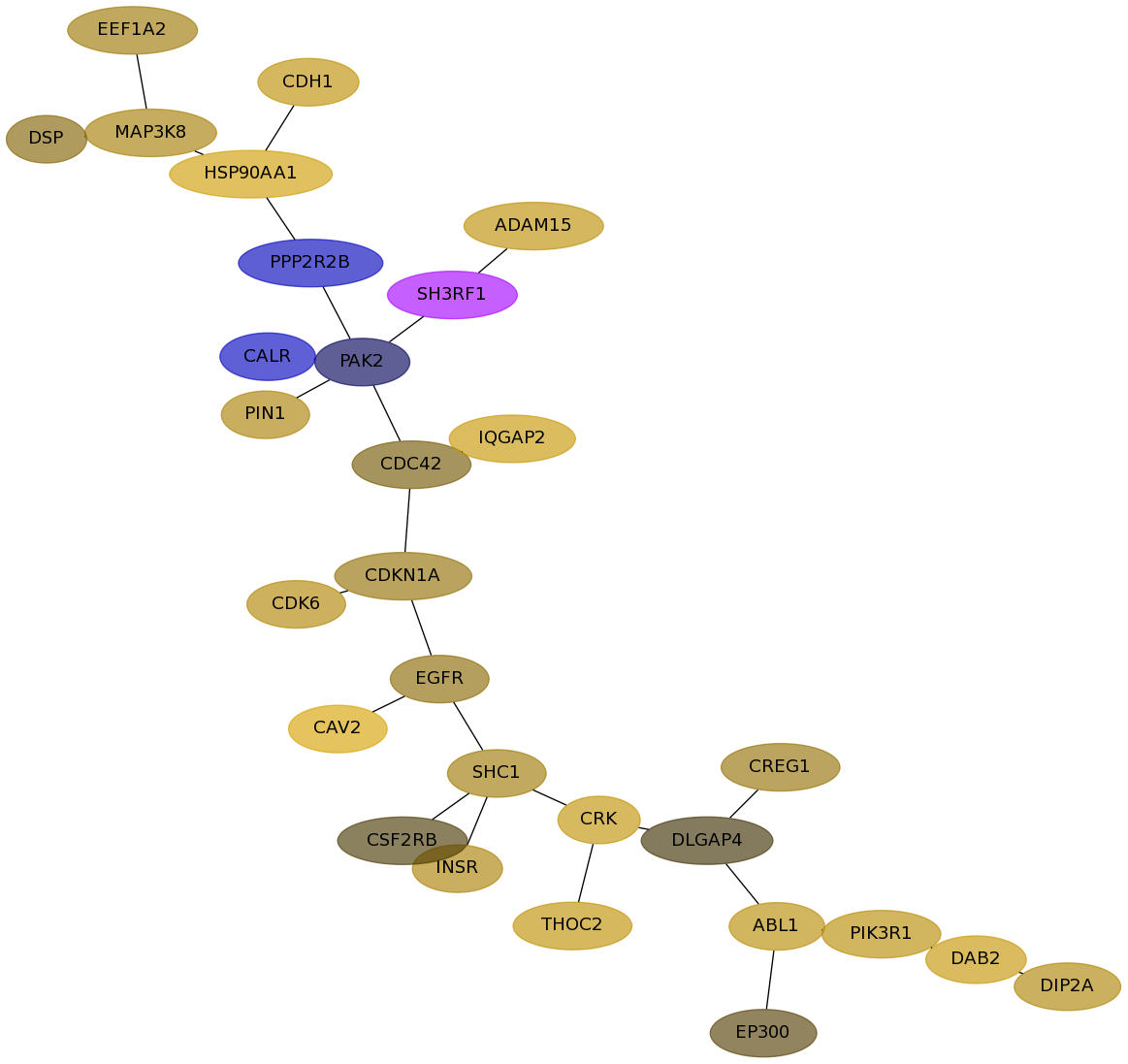
Score for each gene in subnetwork 1158 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| CREG1 |   | 5 | 148 | 691 | 670 | 0.080 | 0.029 | 0.018 | -0.189 | -0.176 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| MAP3K8 |   | 2 | 391 | 691 | 694 | 0.119 | 0.188 | -0.027 | 0.060 | -0.074 |
|---|
| DLGAP4 |   | 5 | 148 | 326 | 330 | 0.010 | -0.029 | 0.180 | -0.201 | -0.043 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| DSP |   | 7 | 100 | 206 | 198 | 0.052 | 0.054 | 0.235 | -0.119 | 0.053 |
|---|
| DIP2A |   | 1 | 652 | 691 | 710 | 0.146 | -0.054 | 0.100 | 0.101 | 0.067 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| CSF2RB |   | 3 | 265 | 691 | 685 | 0.013 | 0.013 | -0.196 | -0.109 | -0.141 |
|---|
| ABL1 |   | 8 | 84 | 326 | 316 | 0.193 | -0.152 | 0.200 | 0.108 | -0.059 |
|---|
| CALR |   | 10 | 67 | 117 | 121 | -0.216 | -0.105 | 0.002 | -0.061 | -0.075 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| SHC1 |   | 4 | 198 | 691 | 671 | 0.102 | -0.090 | -0.041 | -0.005 | -0.138 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| SH3RF1 |   | 14 | 42 | 117 | 120 | undef | 0.144 | 0.238 | undef | undef |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| EEF1A2 |   | 9 | 77 | 182 | 179 | 0.098 | 0.032 | 0.220 | -0.119 | 0.102 |
|---|
| ADAM15 |   | 13 | 47 | 492 | 474 | 0.205 | -0.048 | -0.078 | -0.208 | -0.048 |
|---|
| HSP90AA1 |   | 10 | 67 | 1 | 16 | 0.321 | -0.143 | 0.104 | 0.123 | -0.054 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
GO Enrichment output for subnetwork 1158 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.199E-09 | 9.657E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.178E-07 | 2.505E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 2.376E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 1.782E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 2.845E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 4.139E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.08E-06 | 3.548E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.58E-06 | 7.417E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.677E-06 | 9.397E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 5.045E-06 | 1.16E-03 |
|---|
| microspike assembly | GO:0030035 |  | 6.711E-06 | 1.403E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.165E-09 | 5.289E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.306E-07 | 1.596E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.448E-07 | 2.808E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.448E-07 | 2.106E-04 |
|---|
| interaction with host | GO:0051701 |  | 5.326E-07 | 2.602E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.619E-07 | 3.916E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 9.619E-07 | 3.357E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.44E-06 | 4.398E-04 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 1.918E-06 | 5.205E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.818E-06 | 6.884E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.75E-06 | 8.329E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.865E-09 | 4.486E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.125E-07 | 1.353E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.727E-07 | 2.989E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.727E-07 | 2.242E-04 |
|---|
| interaction with host | GO:0051701 |  | 5.883E-07 | 2.831E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.509E-07 | 2.61E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.509E-07 | 2.237E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.039E-06 | 3.126E-04 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 2.117E-06 | 5.66E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.045E-06 | 7.325E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 4.052E-06 | 8.862E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.199E-09 | 9.657E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.178E-07 | 2.505E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 2.376E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 1.782E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 2.845E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 4.139E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.08E-06 | 3.548E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.58E-06 | 7.417E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.677E-06 | 9.397E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 5.045E-06 | 1.16E-03 |
|---|
| microspike assembly | GO:0030035 |  | 6.711E-06 | 1.403E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.199E-09 | 9.657E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.178E-07 | 2.505E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 2.376E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 1.782E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 2.845E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 4.139E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.08E-06 | 3.548E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.58E-06 | 7.417E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.677E-06 | 9.397E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 5.045E-06 | 1.16E-03 |
|---|
| microspike assembly | GO:0030035 |  | 6.711E-06 | 1.403E-03 |
|---|


