Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1141
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3340 | 1.464e-03 | 2.500e-05 | 1.603e-01 | 5.867e-09 |
|---|
| IPC | 0.3398 | 3.505e-01 | 1.799e-01 | 9.360e-01 | 5.902e-02 |
|---|
| Loi | 0.4756 | 5.700e-05 | 0.000e+00 | 5.382e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5975 | 1.000e-06 | 0.000e+00 | 1.180e-01 | 0.000e+00 |
|---|
| Wang | 0.2576 | 7.564e-03 | 6.588e-02 | 2.288e-01 | 1.140e-04 |
|---|
Expression data for subnetwork 1141 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
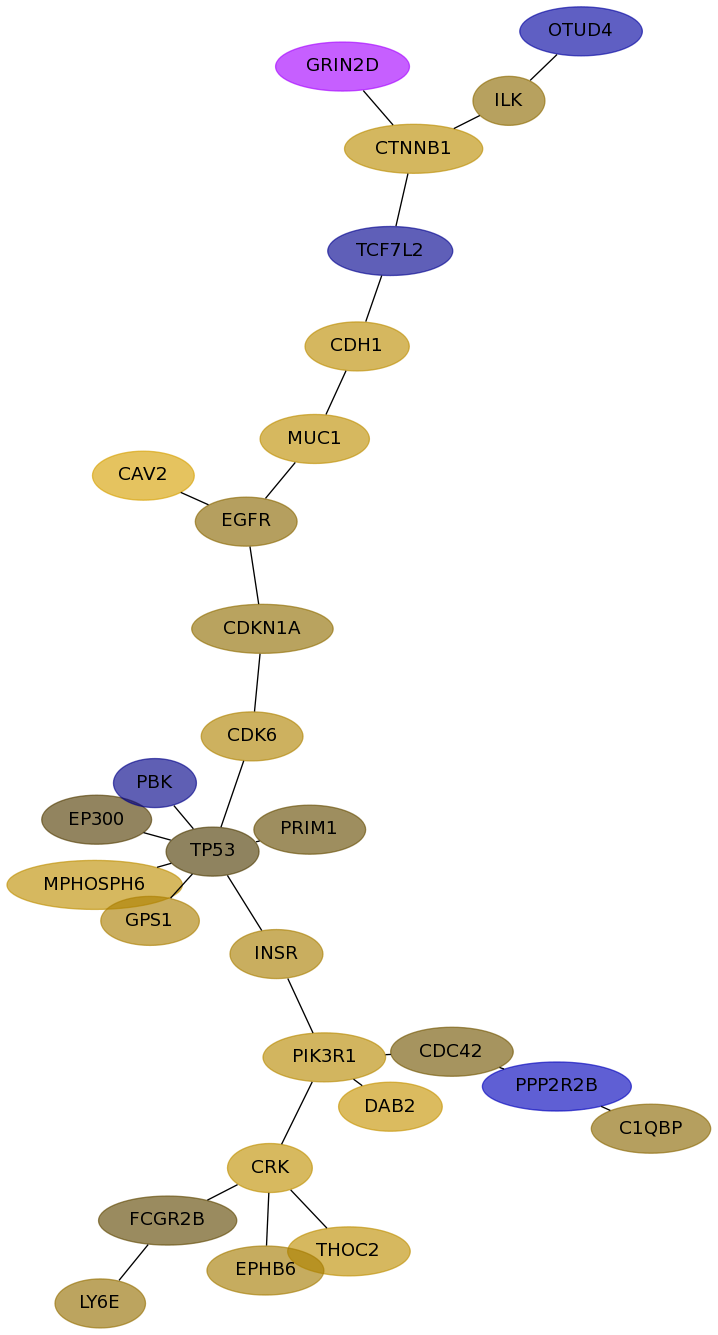
Score for each gene in subnetwork 1141 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| FCGR2B |   | 1 | 652 | 900 | 912 | 0.023 | 0.253 | -0.024 | 0.051 | -0.002 |
|---|
| PBK |   | 1 | 652 | 900 | 912 | -0.062 | 0.163 | -0.164 | 0.107 | -0.095 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| LY6E |   | 1 | 652 | 900 | 912 | 0.081 | 0.132 | -0.173 | -0.063 | -0.226 |
|---|
| ILK |   | 6 | 120 | 33 | 40 | 0.069 | -0.020 | 0.195 | 0.076 | 0.086 |
|---|
| C1QBP |   | 4 | 198 | 552 | 547 | 0.064 | 0.134 | -0.072 | 0.080 | -0.006 |
|---|
| TCF7L2 |   | 17 | 30 | 271 | 264 | -0.071 | -0.021 | 0.143 | 0.280 | 0.101 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| PRIM1 |   | 3 | 265 | 900 | 892 | 0.027 | 0.153 | -0.219 | 0.059 | 0.091 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CTNNB1 |   | 33 | 14 | 33 | 29 | 0.202 | -0.113 | 0.187 | 0.159 | 0.239 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| GRIN2D |   | 18 | 26 | 33 | 32 | undef | 0.058 | 0.155 | 0.000 | undef |
|---|
| OTUD4 |   | 2 | 391 | 900 | 897 | -0.107 | -0.204 | -0.012 | 0.227 | -0.092 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| EPHB6 |   | 7 | 100 | 419 | 407 | 0.122 | 0.008 | 0.042 | -0.219 | -0.059 |
|---|
| GPS1 |   | 3 | 265 | 900 | 892 | 0.137 | -0.239 | -0.126 | 0.057 | -0.145 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| MPHOSPH6 |   | 8 | 84 | 756 | 726 | 0.217 | -0.084 | 0.175 | -0.269 | 0.146 |
|---|
GO Enrichment output for subnetwork 1141 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.271E-07 | 7.524E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.271E-07 | 3.762E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.732E-07 | 3.628E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.527E-07 | 3.753E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 7.11E-07 | 3.27E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.14E-06 | 4.369E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.14E-06 | 3.745E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.819E-06 | 5.23E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.191E-06 | 5.6E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.722E-06 | 6.262E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.648E-06 | 7.628E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 5.299E-11 | 1.294E-07 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.652E-07 | 3.239E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 3.562E-07 | 2.9E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.562E-07 | 2.175E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.562E-07 | 1.74E-04 |
|---|
| interaction with host | GO:0051701 |  | 5.56E-07 | 2.264E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 8.944E-07 | 3.122E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.935E-07 | 3.034E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 9.935E-07 | 2.697E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 9.935E-07 | 2.427E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.487E-06 | 3.303E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.204E-11 | 1.012E-07 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.899E-07 | 3.488E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 4.468E-07 | 3.584E-04 |
|---|
| nuclear migration | GO:0007097 |  | 4.468E-07 | 2.688E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.468E-07 | 2.15E-04 |
|---|
| interaction with host | GO:0051701 |  | 7.487E-07 | 3.002E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 7.803E-07 | 2.682E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 7.803E-07 | 2.347E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 1.034E-06 | 2.765E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 1.246E-06 | 2.998E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.246E-06 | 2.725E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.271E-07 | 7.524E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.271E-07 | 3.762E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.732E-07 | 3.628E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.527E-07 | 3.753E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 7.11E-07 | 3.27E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.14E-06 | 4.369E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.14E-06 | 3.745E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.819E-06 | 5.23E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.191E-06 | 5.6E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.722E-06 | 6.262E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.648E-06 | 7.628E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 3.271E-07 | 7.524E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.271E-07 | 3.762E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.732E-07 | 3.628E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.527E-07 | 3.753E-04 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 7.11E-07 | 3.27E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.14E-06 | 4.369E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.14E-06 | 3.745E-04 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.819E-06 | 5.23E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.191E-06 | 5.6E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.722E-06 | 6.262E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.648E-06 | 7.628E-04 |
|---|


