Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1129
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3329 | 1.537e-03 | 2.700e-05 | 1.637e-01 | 6.794e-09 |
|---|
| IPC | 0.3399 | 3.504e-01 | 1.798e-01 | 9.360e-01 | 5.897e-02 |
|---|
| Loi | 0.4737 | 6.200e-05 | 0.000e+00 | 5.479e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5989 | 1.000e-06 | 0.000e+00 | 1.151e-01 | 0.000e+00 |
|---|
| Wang | 0.2582 | 7.377e-03 | 6.486e-02 | 2.262e-01 | 1.082e-04 |
|---|
Expression data for subnetwork 1129 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
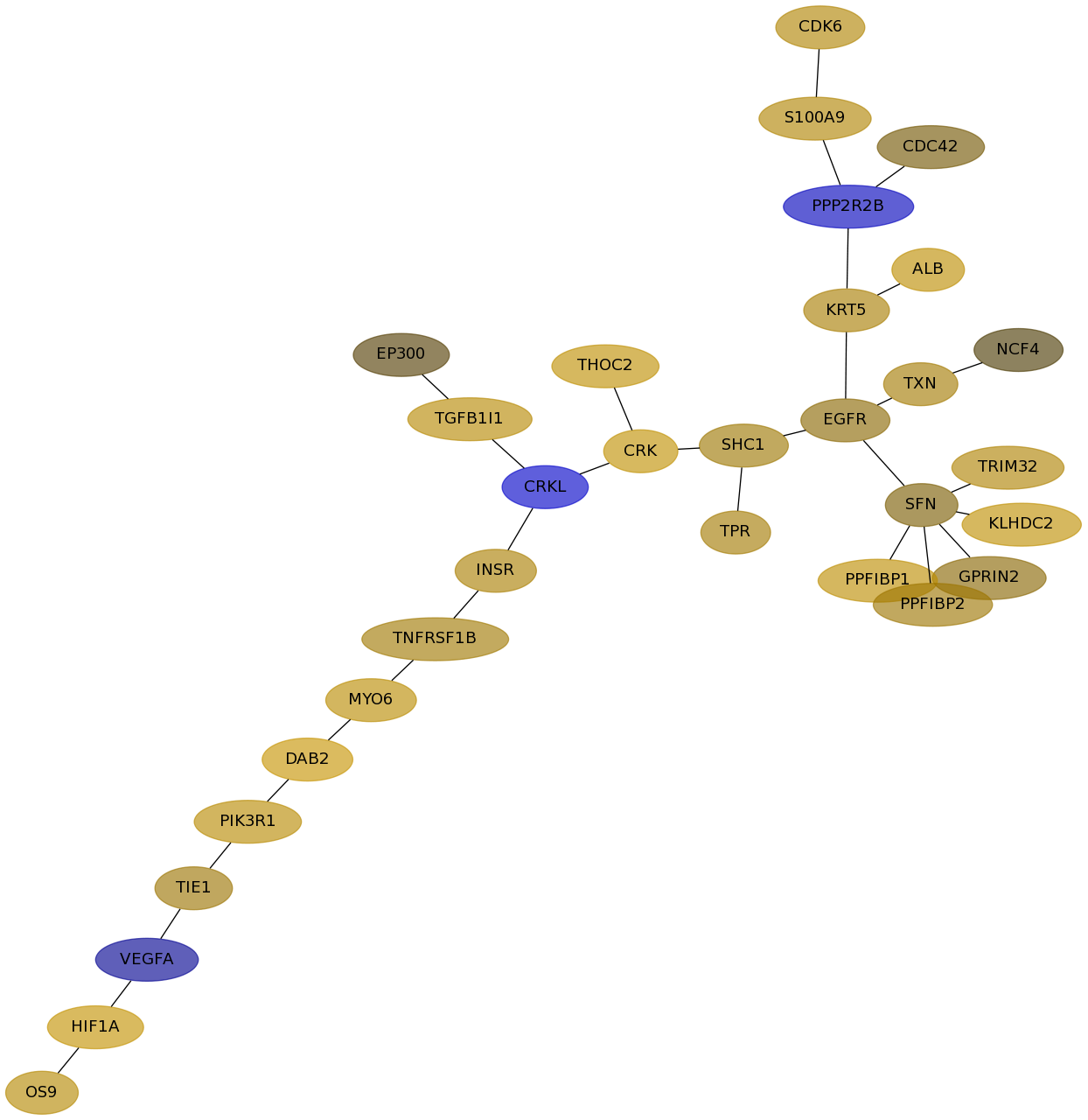
Score for each gene in subnetwork 1129 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| NCF4 |   | 1 | 652 | 923 | 939 | 0.014 | 0.145 | -0.249 | -0.142 | -0.088 |
|---|
| KRT5 |   | 5 | 148 | 923 | 898 | 0.129 | -0.125 | 0.046 | 0.064 | -0.190 |
|---|
| TXN |   | 5 | 148 | 657 | 634 | 0.113 | 0.124 | 0.012 | 0.014 | -0.006 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PPFIBP2 |   | 7 | 100 | 606 | 576 | 0.098 | 0.188 | 0.320 | -0.112 | 0.013 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| OS9 |   | 1 | 652 | 923 | 939 | 0.175 | -0.118 | 0.158 | -0.160 | 0.068 |
|---|
| TNFRSF1B |   | 12 | 55 | 563 | 544 | 0.109 | 0.024 | -0.187 | -0.096 | -0.106 |
|---|
| CRKL |   | 7 | 100 | 326 | 317 | -0.271 | -0.023 | 0.056 | 0.271 | -0.008 |
|---|
| TPR |   | 10 | 67 | 148 | 152 | 0.118 | -0.130 | 0.120 | -0.036 | 0.156 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| DAB2 |   | 13 | 47 | 1 | 15 | 0.258 | -0.175 | 0.067 | -0.022 | 0.071 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| SHC1 |   | 4 | 198 | 691 | 671 | 0.102 | -0.090 | -0.041 | -0.005 | -0.138 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| ALB |   | 7 | 100 | 57 | 60 | 0.207 | -0.122 | 0.218 | 0.080 | 0.038 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| GPRIN2 |   | 1 | 652 | 923 | 939 | 0.061 | 0.084 | 0.078 | 0.012 | 0.114 |
|---|
| PPFIBP1 |   | 2 | 391 | 859 | 860 | 0.212 | -0.073 | 0.207 | -0.072 | -0.016 |
|---|
| TIE1 |   | 6 | 120 | 148 | 156 | 0.094 | -0.108 | 0.005 | 0.141 | 0.252 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| TRIM32 |   | 1 | 652 | 923 | 939 | 0.151 | 0.029 | 0.097 | 0.283 | 0.067 |
|---|
| MYO6 |   | 3 | 265 | 923 | 909 | 0.197 | -0.011 | 0.139 | -0.128 | -0.031 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| CDK6 |   | 25 | 19 | 1 | 10 | 0.156 | -0.229 | 0.107 | -0.028 | 0.104 |
|---|
| SFN |   | 3 | 265 | 806 | 794 | 0.044 | 0.141 | 0.270 | 0.023 | 0.101 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
| KLHDC2 |   | 2 | 391 | 923 | 915 | 0.219 | 0.082 | 0.065 | -0.030 | 0.079 |
|---|
GO Enrichment output for subnetwork 1129 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.437E-07 | 1.021E-03 |
|---|
| nuclear migration | GO:0007097 |  | 4.437E-07 | 5.103E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.437E-07 | 3.402E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.852E-07 | 5.09E-04 |
|---|
| interaction with symbiont | GO:0051702 |  | 8.852E-07 | 4.072E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 8.852E-07 | 3.393E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.069E-06 | 3.512E-04 |
|---|
| interaction with host | GO:0051701 |  | 1.288E-06 | 3.703E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.545E-06 | 3.949E-04 |
|---|
| protein heterooligomerization | GO:0051291 |  | 1.545E-06 | 3.554E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.545E-06 | 3.231E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interaction with host | GO:0051701 |  | 6.938E-09 | 1.695E-05 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 3.564E-08 | 4.353E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.962E-07 | 1.598E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.917E-07 | 2.392E-04 |
|---|
| interaction with symbiont | GO:0051702 |  | 3.917E-07 | 1.914E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.917E-07 | 1.595E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.573E-07 | 1.945E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 9.895E-07 | 3.022E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.092E-06 | 2.965E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.092E-06 | 2.669E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 1.092E-06 | 2.426E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interaction with host | GO:0051701 |  | 9.089E-09 | 2.187E-05 |
|---|
| symbiosis. encompassing mutualism through parasitism | GO:0044403 |  | 4.664E-08 | 5.61E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.319E-07 | 1.86E-04 |
|---|
| nuclear migration | GO:0007097 |  | 4.628E-07 | 2.783E-04 |
|---|
| interaction with symbiont | GO:0051702 |  | 4.628E-07 | 2.227E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.628E-07 | 1.856E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 6.661E-07 | 2.289E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 8.081E-07 | 2.43E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.081E-07 | 2.16E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.194E-06 | 2.872E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.29E-06 | 2.822E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.437E-07 | 1.021E-03 |
|---|
| nuclear migration | GO:0007097 |  | 4.437E-07 | 5.103E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.437E-07 | 3.402E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.852E-07 | 5.09E-04 |
|---|
| interaction with symbiont | GO:0051702 |  | 8.852E-07 | 4.072E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 8.852E-07 | 3.393E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.069E-06 | 3.512E-04 |
|---|
| interaction with host | GO:0051701 |  | 1.288E-06 | 3.703E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.545E-06 | 3.949E-04 |
|---|
| protein heterooligomerization | GO:0051291 |  | 1.545E-06 | 3.554E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.545E-06 | 3.231E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 4.437E-07 | 1.021E-03 |
|---|
| nuclear migration | GO:0007097 |  | 4.437E-07 | 5.103E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.437E-07 | 3.402E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 8.852E-07 | 5.09E-04 |
|---|
| interaction with symbiont | GO:0051702 |  | 8.852E-07 | 4.072E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 8.852E-07 | 3.393E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.069E-06 | 3.512E-04 |
|---|
| interaction with host | GO:0051701 |  | 1.288E-06 | 3.703E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.545E-06 | 3.949E-04 |
|---|
| protein heterooligomerization | GO:0051291 |  | 1.545E-06 | 3.554E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.545E-06 | 3.231E-04 |
|---|


