Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1125
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3328 | 1.548e-03 | 2.700e-05 | 1.642e-01 | 6.862e-09 |
|---|
| IPC | 0.3412 | 3.482e-01 | 1.779e-01 | 9.355e-01 | 5.795e-02 |
|---|
| Loi | 0.4733 | 6.400e-05 | 0.000e+00 | 5.501e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5991 | 1.000e-06 | 0.000e+00 | 1.147e-01 | 0.000e+00 |
|---|
| Wang | 0.2580 | 7.436e-03 | 6.518e-02 | 2.270e-01 | 1.100e-04 |
|---|
Expression data for subnetwork 1125 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
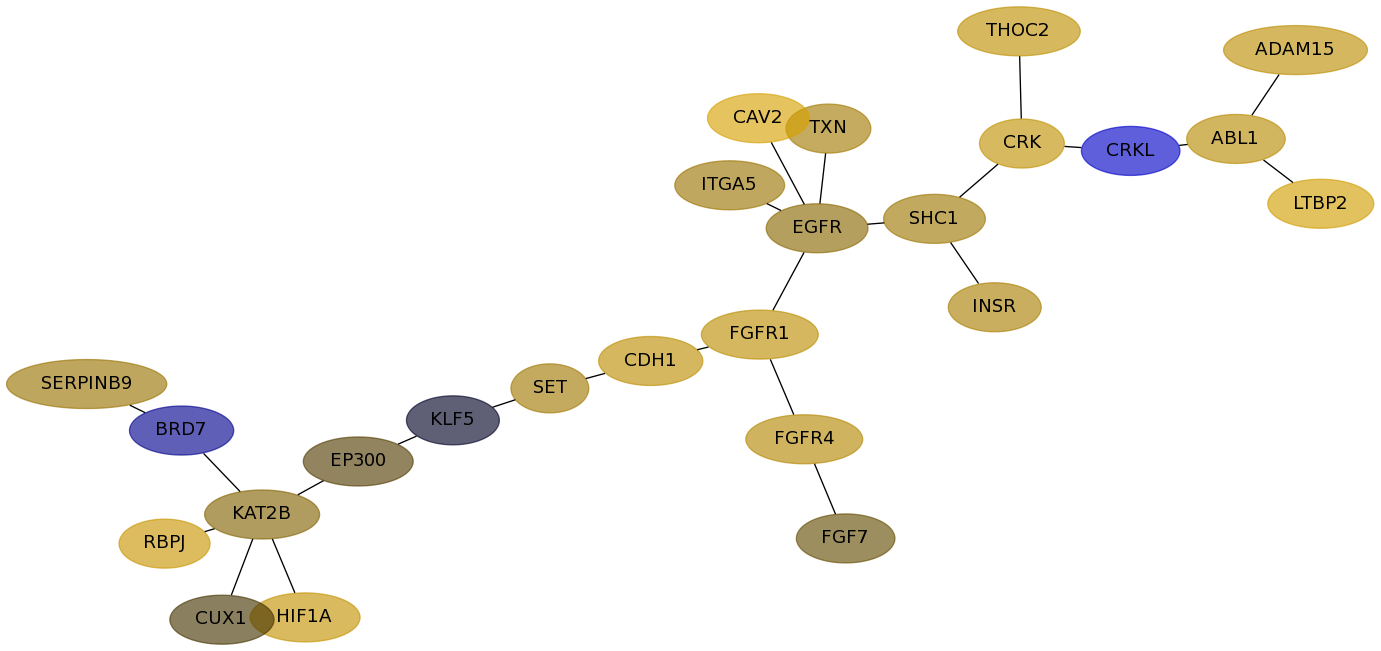
Score for each gene in subnetwork 1125 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| TXN |   | 5 | 148 | 657 | 634 | 0.113 | 0.124 | 0.012 | 0.014 | -0.006 |
|---|
| KAT2B |   | 1 | 652 | 906 | 916 | 0.055 | 0.102 | -0.157 | -0.091 | -0.087 |
|---|
| SERPINB9 |   | 2 | 391 | 647 | 646 | 0.091 | -0.094 | -0.073 | -0.025 | 0.002 |
|---|
| CUX1 |   | 8 | 84 | 271 | 266 | 0.012 | -0.042 | 0.256 | -0.121 | 0.040 |
|---|
| ITGA5 |   | 5 | 148 | 339 | 337 | 0.093 | 0.031 | 0.192 | 0.125 | 0.202 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| CRKL |   | 7 | 100 | 326 | 317 | -0.271 | -0.023 | 0.056 | 0.271 | -0.008 |
|---|
| ABL1 |   | 8 | 84 | 326 | 316 | 0.193 | -0.152 | 0.200 | 0.108 | -0.059 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| SHC1 |   | 4 | 198 | 691 | 671 | 0.102 | -0.090 | -0.041 | -0.005 | -0.138 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CRK |   | 27 | 17 | 206 | 193 | 0.232 | -0.116 | 0.106 | 0.090 | 0.017 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| THOC2 |   | 11 | 59 | 419 | 406 | 0.217 | -0.094 | 0.119 | 0.068 | 0.072 |
|---|
| SET |   | 3 | 265 | 606 | 599 | 0.108 | 0.103 | 0.250 | 0.196 | -0.128 |
|---|
| FGF7 |   | 8 | 84 | 398 | 384 | 0.026 | -0.094 | 0.237 | 0.070 | 0.083 |
|---|
| ADAM15 |   | 13 | 47 | 492 | 474 | 0.205 | -0.048 | -0.078 | -0.208 | -0.048 |
|---|
| BRD7 |   | 4 | 198 | 906 | 894 | -0.068 | -0.090 | 0.019 | -0.188 | -0.086 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| RBPJ |   | 1 | 652 | 906 | 916 | 0.273 | -0.061 | 0.049 | -0.029 | 0.125 |
|---|
| LTBP2 |   | 1 | 652 | 906 | 916 | 0.339 | -0.120 | 0.222 | 0.081 | 0.159 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
GO Enrichment output for subnetwork 1125 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 4.109E-09 | 9.45E-06 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 6.357E-09 | 7.311E-06 |
|---|
| regulation of histone modification | GO:0031056 |  | 7.38E-09 | 5.658E-06 |
|---|
| positive regulation of oxidoreductase activity | GO:0051353 |  | 7.895E-08 | 4.54E-05 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 1.371E-07 | 6.306E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.193E-07 | 8.405E-05 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.534E-07 | 8.325E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.773E-07 | 7.972E-05 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 4.169E-07 | 1.065E-04 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 4.376E-07 | 1.007E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 4.376E-07 | 9.15E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| lung development | GO:0030324 |  | 1.494E-10 | 3.649E-07 |
|---|
| respiratory tube development | GO:0030323 |  | 2.029E-10 | 2.478E-07 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.371E-10 | 1.931E-07 |
|---|
| respiratory system development | GO:0060541 |  | 2.47E-10 | 1.509E-07 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 1.001E-09 | 4.889E-07 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 1.001E-09 | 4.074E-07 |
|---|
| nucleosome disassembly | GO:0006337 |  | 1.342E-09 | 4.683E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.61E-09 | 4.916E-07 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 1.61E-09 | 4.369E-07 |
|---|
| regulation of histone modification | GO:0031056 |  | 2.411E-09 | 5.891E-07 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 4.46E-09 | 9.905E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| lung development | GO:0030324 |  | 1.528E-10 | 3.678E-07 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.005E-10 | 2.413E-07 |
|---|
| respiratory tube development | GO:0030323 |  | 2.096E-10 | 1.681E-07 |
|---|
| respiratory system development | GO:0060541 |  | 2.567E-10 | 1.544E-07 |
|---|
| inner ear receptor cell differentiation | GO:0060113 |  | 7.07E-10 | 3.402E-07 |
|---|
| intra-Golgi vesicle-mediated transport | GO:0006891 |  | 9.263E-10 | 3.714E-07 |
|---|
| mechanoreceptor differentiation | GO:0042490 |  | 1.197E-09 | 4.113E-07 |
|---|
| nucleosome disassembly | GO:0006337 |  | 1.555E-09 | 4.677E-07 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 1.925E-09 | 5.146E-07 |
|---|
| regulation of histone modification | GO:0031056 |  | 2.794E-09 | 6.723E-07 |
|---|
| positive regulation of oxidoreductase activity | GO:0051353 |  | 2.997E-08 | 6.555E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 4.109E-09 | 9.45E-06 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 6.357E-09 | 7.311E-06 |
|---|
| regulation of histone modification | GO:0031056 |  | 7.38E-09 | 5.658E-06 |
|---|
| positive regulation of oxidoreductase activity | GO:0051353 |  | 7.895E-08 | 4.54E-05 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 1.371E-07 | 6.306E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.193E-07 | 8.405E-05 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.534E-07 | 8.325E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.773E-07 | 7.972E-05 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 4.169E-07 | 1.065E-04 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 4.376E-07 | 1.007E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 4.376E-07 | 9.15E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 4.109E-09 | 9.45E-06 |
|---|
| regulation of oxidoreductase activity | GO:0051341 |  | 6.357E-09 | 7.311E-06 |
|---|
| regulation of histone modification | GO:0031056 |  | 7.38E-09 | 5.658E-06 |
|---|
| positive regulation of oxidoreductase activity | GO:0051353 |  | 7.895E-08 | 4.54E-05 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 1.371E-07 | 6.306E-05 |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 2.193E-07 | 8.405E-05 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 2.534E-07 | 8.325E-05 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.773E-07 | 7.972E-05 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 4.169E-07 | 1.065E-04 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 4.376E-07 | 1.007E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 4.376E-07 | 9.15E-05 |
|---|


