Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1123
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3325 | 1.569e-03 | 2.800e-05 | 1.651e-01 | 7.255e-09 |
|---|
| IPC | 0.3423 | 3.464e-01 | 1.763e-01 | 9.352e-01 | 5.711e-02 |
|---|
| Loi | 0.4729 | 6.500e-05 | 0.000e+00 | 5.522e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5990 | 1.000e-06 | 0.000e+00 | 1.148e-01 | 0.000e+00 |
|---|
| Wang | 0.2581 | 7.407e-03 | 6.502e-02 | 2.266e-01 | 1.091e-04 |
|---|
Expression data for subnetwork 1123 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
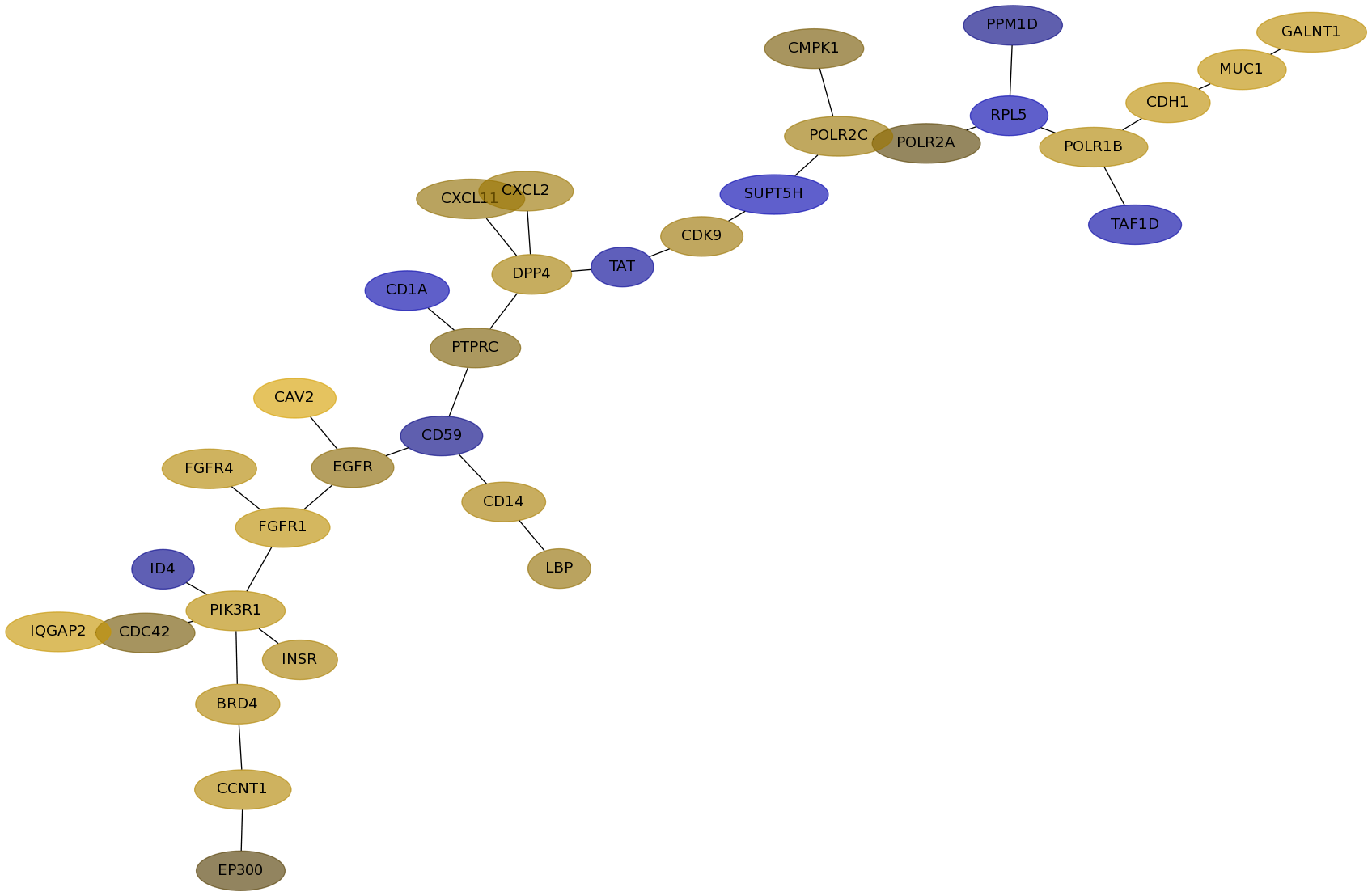
Score for each gene in subnetwork 1123 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| PPM1D |   | 2 | 391 | 889 | 891 | -0.050 | 0.105 | 0.121 | 0.125 | 0.166 |
|---|
| ID4 |   | 13 | 47 | 444 | 430 | -0.060 | -0.179 | 0.137 | 0.241 | 0.036 |
|---|
| CXCL2 |   | 1 | 652 | 889 | 901 | 0.095 | 0.039 | -0.026 | 0.108 | -0.079 |
|---|
| RPL5 |   | 4 | 198 | 889 | 873 | -0.145 | -0.000 | 0.128 | 0.220 | -0.110 |
|---|
| CD1A |   | 1 | 652 | 889 | 901 | -0.134 | -0.098 | -0.005 | 0.040 | 0.002 |
|---|
| IQGAP2 |   | 16 | 34 | 117 | 119 | 0.264 | -0.170 | -0.051 | -0.112 | 0.049 |
|---|
| CD59 |   | 1 | 652 | 889 | 901 | -0.051 | -0.032 | 0.228 | 0.013 | -0.010 |
|---|
| GALNT1 |   | 3 | 265 | 889 | 874 | 0.200 | 0.133 | -0.061 | -0.181 | 0.234 |
|---|
| CDH1 |   | 54 | 5 | 1 | 4 | 0.211 | -0.227 | 0.187 | -0.236 | -0.024 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| POLR1B |   | 11 | 59 | 129 | 126 | 0.157 | 0.062 | -0.036 | -0.079 | 0.139 |
|---|
| CMPK1 |   | 2 | 391 | 756 | 756 | 0.039 | -0.054 | 0.193 | -0.038 | 0.096 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| TAT |   | 1 | 652 | 889 | 901 | -0.080 | -0.113 | 0.030 | -0.182 | -0.074 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| TAF1D |   | 5 | 148 | 57 | 63 | -0.110 | 0.141 | 0.113 | 0.041 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CD14 |   | 1 | 652 | 889 | 901 | 0.128 | 0.183 | -0.144 | 0.050 | 0.013 |
|---|
| SUPT5H |   | 9 | 77 | 1 | 18 | -0.150 | -0.085 | 0.007 | -0.298 | -0.068 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| POLR2A |   | 5 | 148 | 271 | 267 | 0.019 | -0.107 | 0.247 | 0.091 | 0.141 |
|---|
| POLR2C |   | 2 | 391 | 756 | 756 | 0.097 | -0.130 | 0.002 | -0.204 | 0.168 |
|---|
| PTPRC |   | 5 | 148 | 715 | 697 | 0.043 | -0.031 | -0.204 | -0.050 | -0.068 |
|---|
| MUC1 |   | 39 | 11 | 1 | 7 | 0.215 | 0.063 | 0.214 | -0.190 | -0.132 |
|---|
| CXCL11 |   | 1 | 652 | 889 | 901 | 0.074 | 0.256 | -0.245 | -0.143 | -0.064 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| CDK9 |   | 4 | 198 | 129 | 130 | 0.095 | -0.120 | 0.179 | 0.182 | -0.135 |
|---|
| DPP4 |   | 1 | 652 | 889 | 901 | 0.123 | -0.072 | -0.192 | 0.012 | 0.166 |
|---|
| LBP |   | 1 | 652 | 889 | 901 | 0.078 | -0.154 | 0.135 | -0.226 | -0.019 |
|---|
GO Enrichment output for subnetwork 1123 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 1.206E-07 | 2.774E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.382E-07 | 7.34E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.382E-07 | 4.893E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 6.382E-07 | 3.67E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.152E-06 | 5.299E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.273E-06 | 4.879E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.221E-06 | 7.297E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.221E-06 | 6.385E-04 |
|---|
| nucleus localization | GO:0051647 |  | 5.299E-06 | 1.354E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 7.548E-06 | 1.736E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.035E-05 | 2.164E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 4.66E-08 | 1.138E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.832E-07 | 4.68E-04 |
|---|
| nuclear migration | GO:0007097 |  | 4.697E-07 | 3.825E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 4.697E-07 | 2.868E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.309E-06 | 6.398E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.309E-06 | 5.332E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 1.309E-06 | 4.57E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.96E-06 | 5.986E-04 |
|---|
| nucleus localization | GO:0051647 |  | 3.834E-06 | 1.041E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.101E-06 | 1.246E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.56E-05 | 3.465E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 5.461E-08 | 1.314E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.641E-07 | 4.38E-04 |
|---|
| nuclear migration | GO:0007097 |  | 5.302E-07 | 4.252E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 5.302E-07 | 3.189E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 9.258E-07 | 4.455E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 9.258E-07 | 3.713E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 9.258E-07 | 3.182E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.478E-06 | 4.445E-04 |
|---|
| nucleus localization | GO:0051647 |  | 4.327E-06 | 1.157E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 5.756E-06 | 1.385E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.452E-05 | 3.177E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 1.206E-07 | 2.774E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.382E-07 | 7.34E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.382E-07 | 4.893E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 6.382E-07 | 3.67E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.152E-06 | 5.299E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.273E-06 | 4.879E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.221E-06 | 7.297E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.221E-06 | 6.385E-04 |
|---|
| nucleus localization | GO:0051647 |  | 5.299E-06 | 1.354E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 7.548E-06 | 1.736E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.035E-05 | 2.164E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ribosomal large subunit biogenesis | GO:0042273 |  | 1.206E-07 | 2.774E-04 |
|---|
| nuclear migration | GO:0007097 |  | 6.382E-07 | 7.34E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 6.382E-07 | 4.893E-04 |
|---|
| regulation of respiratory burst | GO:0060263 |  | 6.382E-07 | 3.67E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.152E-06 | 5.299E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 1.273E-06 | 4.879E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.221E-06 | 7.297E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 2.221E-06 | 6.385E-04 |
|---|
| nucleus localization | GO:0051647 |  | 5.299E-06 | 1.354E-03 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 7.548E-06 | 1.736E-03 |
|---|
| filopodium assembly | GO:0046847 |  | 1.035E-05 | 2.164E-03 |
|---|


