Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1119
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3329 | 1.538e-03 | 2.700e-05 | 1.638e-01 | 6.801e-09 |
|---|
| IPC | 0.3440 | 3.435e-01 | 1.738e-01 | 9.346e-01 | 5.581e-02 |
|---|
| Loi | 0.4728 | 6.500e-05 | 0.000e+00 | 5.524e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5986 | 1.000e-06 | 0.000e+00 | 1.156e-01 | 0.000e+00 |
|---|
| Wang | 0.2575 | 7.586e-03 | 6.600e-02 | 2.291e-01 | 1.147e-04 |
|---|
Expression data for subnetwork 1119 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
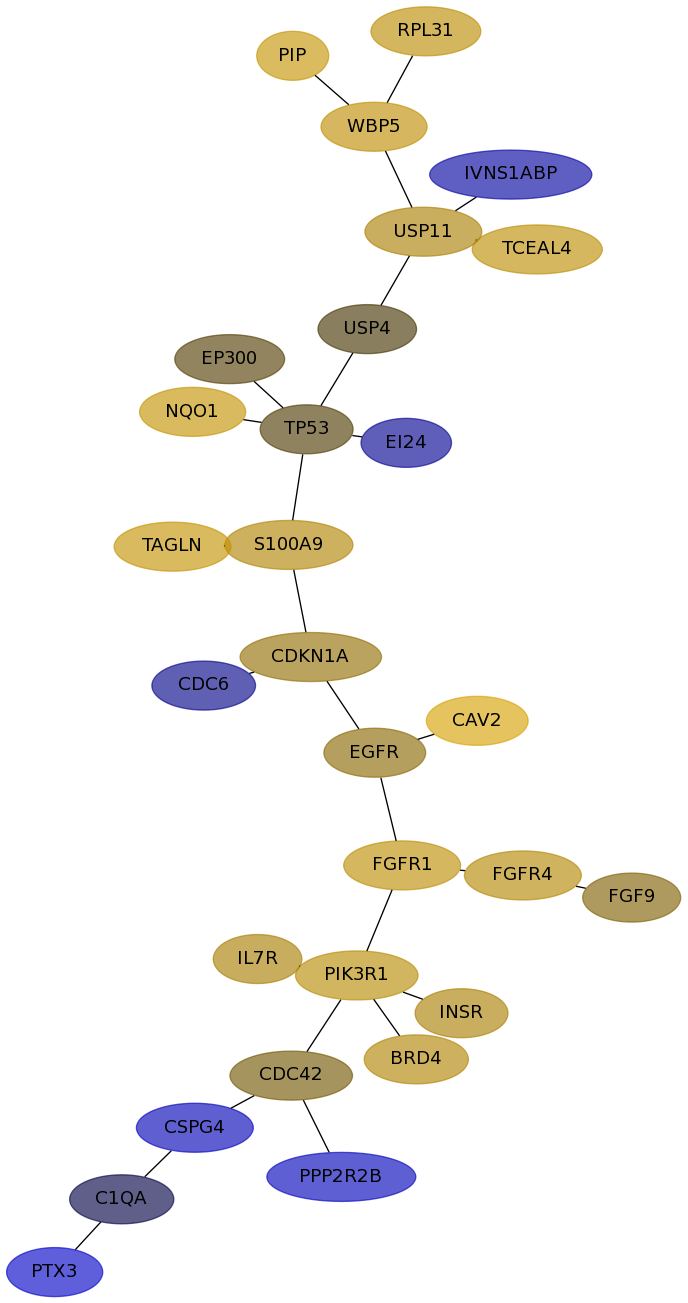
Score for each gene in subnetwork 1119 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| FGF9 |   | 5 | 148 | 262 | 265 | 0.050 | -0.096 | 0.087 | 0.014 | 0.174 |
|---|
| USP4 |   | 6 | 120 | 823 | 803 | 0.012 | -0.093 | 0.147 | 0.097 | 0.054 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| NQO1 |   | 6 | 120 | 536 | 535 | 0.243 | -0.115 | 0.069 | -0.189 | 0.118 |
|---|
| IVNS1ABP |   | 1 | 652 | 823 | 838 | -0.109 | 0.058 | 0.095 | -0.045 | -0.047 |
|---|
| RPL31 |   | 2 | 391 | 823 | 817 | 0.200 | -0.027 | -0.118 | 0.113 | 0.003 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| CDC6 |   | 4 | 198 | 283 | 289 | -0.059 | 0.198 | -0.053 | 0.038 | 0.095 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| PIP |   | 2 | 391 | 823 | 817 | 0.253 | -0.166 | 0.192 | -0.153 | -0.117 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| C1QA |   | 1 | 652 | 823 | 838 | -0.012 | 0.205 | -0.229 | -0.116 | -0.037 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| PTX3 |   | 1 | 652 | 823 | 838 | -0.273 | -0.122 | 0.051 | 0.213 | -0.054 |
|---|
| TCEAL4 |   | 3 | 265 | 552 | 550 | 0.205 | -0.156 | 0.219 | 0.091 | 0.007 |
|---|
| EI24 |   | 8 | 84 | 339 | 336 | -0.074 | 0.051 | 0.078 | 0.107 | 0.070 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| USP11 |   | 5 | 148 | 283 | 287 | 0.134 | -0.144 | 0.106 | 0.121 | -0.044 |
|---|
| S100A9 |   | 18 | 26 | 398 | 382 | 0.154 | 0.117 | 0.043 | -0.110 | -0.045 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| CSPG4 |   | 2 | 391 | 823 | 817 | -0.188 | 0.051 | 0.088 | 0.190 | -0.027 |
|---|
| WBP5 |   | 2 | 391 | 823 | 817 | 0.214 | -0.089 | 0.073 | 0.201 | 0.142 |
|---|
| TAGLN |   | 4 | 198 | 823 | 807 | 0.243 | -0.079 | 0.246 | 0.170 | 0.175 |
|---|
GO Enrichment output for subnetwork 1119 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.199E-09 | 9.657E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.178E-07 | 2.505E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 2.376E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 1.782E-04 |
|---|
| male sex determination | GO:0030238 |  | 6.185E-07 | 2.845E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 2.371E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 3.548E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.08E-06 | 3.105E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.58E-06 | 6.593E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.677E-06 | 8.457E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 5.045E-06 | 1.055E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.141E-09 | 2.788E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 7.854E-08 | 9.593E-05 |
|---|
| nuclear migration | GO:0007097 |  | 2.355E-07 | 1.918E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.355E-07 | 1.438E-04 |
|---|
| male sex determination | GO:0030238 |  | 4.114E-07 | 2.01E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 6.572E-07 | 2.676E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 6.572E-07 | 2.294E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 9.841E-07 | 3.005E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.927E-06 | 5.23E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 2.565E-06 | 6.265E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 3.151E-06 | 6.999E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.176E-09 | 2.829E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 7.799E-08 | 9.383E-05 |
|---|
| nuclear migration | GO:0007097 |  | 2.833E-07 | 2.272E-04 |
|---|
| male sex determination | GO:0030238 |  | 2.833E-07 | 1.704E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.833E-07 | 1.363E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.949E-07 | 1.984E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 4.949E-07 | 1.701E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 7.904E-07 | 2.377E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.316E-06 | 6.193E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.083E-06 | 7.418E-04 |
|---|
| leukocyte differentiation | GO:0002521 |  | 4.01E-06 | 8.77E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.199E-09 | 9.657E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.178E-07 | 2.505E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 2.376E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 1.782E-04 |
|---|
| male sex determination | GO:0030238 |  | 6.185E-07 | 2.845E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 2.371E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 3.548E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.08E-06 | 3.105E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.58E-06 | 6.593E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.677E-06 | 8.457E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 5.045E-06 | 1.055E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.199E-09 | 9.657E-06 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.178E-07 | 2.505E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.099E-07 | 2.376E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.099E-07 | 1.782E-04 |
|---|
| male sex determination | GO:0030238 |  | 6.185E-07 | 2.845E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.185E-07 | 2.371E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.08E-06 | 3.548E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.08E-06 | 3.105E-04 |
|---|
| nucleus localization | GO:0051647 |  | 2.58E-06 | 6.593E-04 |
|---|
| regulation of cell projection assembly | GO:0060491 |  | 3.677E-06 | 8.457E-04 |
|---|
| filopodium assembly | GO:0046847 |  | 5.045E-06 | 1.055E-03 |
|---|


