Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1100
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3351 | 1.389e-03 | 2.300e-05 | 1.567e-01 | 5.005e-09 |
|---|
| IPC | 0.3401 | 3.499e-01 | 1.794e-01 | 9.359e-01 | 5.876e-02 |
|---|
| Loi | 0.4719 | 6.800e-05 | 0.000e+00 | 5.573e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5959 | 1.000e-06 | 0.000e+00 | 1.215e-01 | 0.000e+00 |
|---|
| Wang | 0.2562 | 7.985e-03 | 6.815e-02 | 2.346e-01 | 1.277e-04 |
|---|
Expression data for subnetwork 1100 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
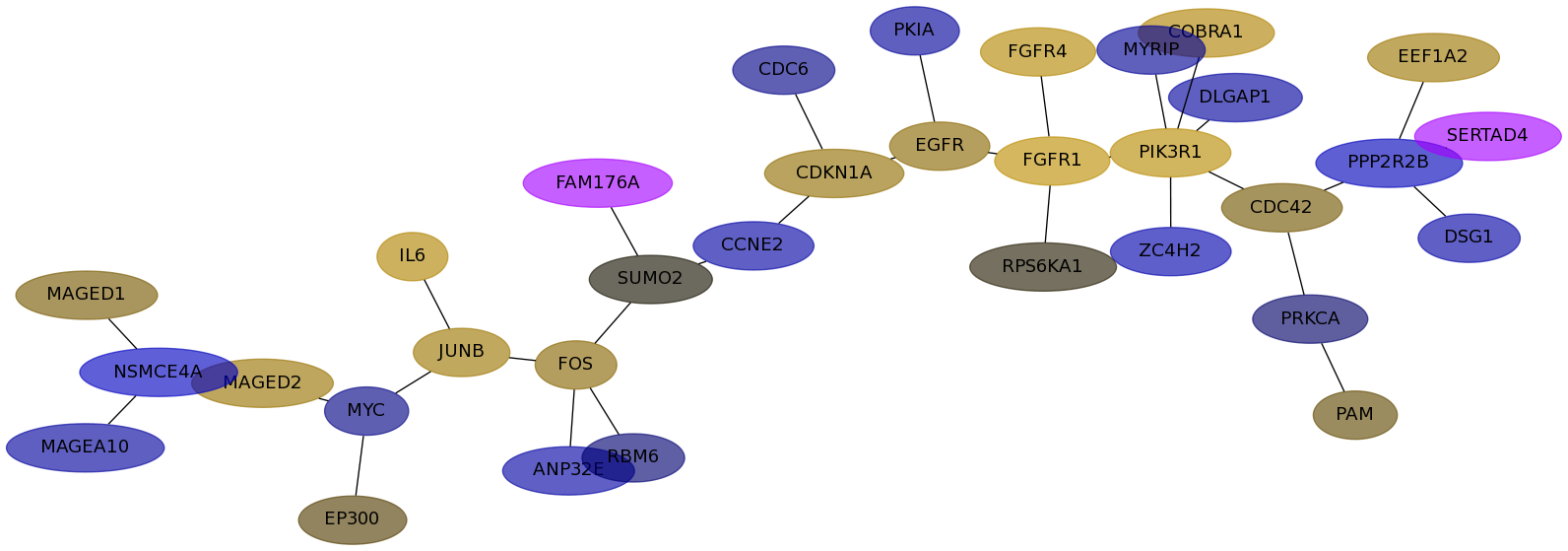
Score for each gene in subnetwork 1100 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| CCNE2 |   | 8 | 84 | 536 | 515 | -0.123 | 0.136 | -0.178 | 0.241 | -0.055 |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| NSMCE4A |   | 3 | 265 | 715 | 705 | -0.228 | 0.177 | -0.116 | 0.290 | -0.091 |
|---|
| RPS6KA1 |   | 8 | 84 | 696 | 669 | 0.005 | -0.208 | 0.084 | -0.322 | -0.071 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| MAGED1 |   | 6 | 120 | 715 | 695 | 0.040 | -0.141 | 0.284 | -0.152 | 0.045 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| PRKCA |   | 5 | 148 | 339 | 337 | -0.029 | -0.016 | -0.088 | 0.174 | 0.005 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| MYC |   | 24 | 21 | 148 | 137 | -0.056 | -0.101 | -0.169 | 0.188 | -0.134 |
|---|
| EEF1A2 |   | 9 | 77 | 182 | 179 | 0.098 | 0.032 | 0.220 | -0.119 | 0.102 |
|---|
| JUNB |   | 5 | 148 | 696 | 672 | 0.101 | 0.057 | 0.086 | 0.065 | -0.015 |
|---|
| RBM6 |   | 10 | 67 | 148 | 152 | -0.036 | -0.082 | 0.265 | -0.004 | 0.111 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| MYRIP |   | 1 | 652 | 1065 | 1073 | -0.084 | -0.190 | 0.007 | 0.232 | -0.017 |
|---|
| COBRA1 |   | 4 | 198 | 206 | 205 | 0.150 | 0.137 | 0.335 | 0.159 | -0.016 |
|---|
| DLGAP1 |   | 2 | 391 | 910 | 899 | -0.104 | 0.103 | 0.078 | 0.102 | -0.159 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| FAM176A |   | 2 | 391 | 1065 | 1059 | undef | 0.054 | 0.217 | undef | undef |
|---|
| SERTAD4 |   | 7 | 100 | 1 | 20 | undef | 0.173 | 0.169 | undef | undef |
|---|
| ZC4H2 |   | 16 | 34 | 339 | 332 | -0.145 | 0.065 | 0.061 | 0.304 | 0.059 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| CDC6 |   | 4 | 198 | 283 | 289 | -0.059 | 0.198 | -0.053 | 0.038 | 0.095 |
|---|
| FOS |   | 20 | 24 | 148 | 138 | 0.060 | -0.137 | 0.129 | 0.185 | 0.234 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| MAGED2 |   | 2 | 391 | 1065 | 1059 | 0.089 | -0.104 | 0.198 | 0.022 | -0.020 |
|---|
| MAGEA10 |   | 1 | 652 | 1065 | 1073 | -0.105 | undef | 0.022 | 0.152 | undef |
|---|
| IL6 |   | 3 | 265 | 715 | 705 | 0.155 | 0.257 | -0.106 | 0.226 | 0.035 |
|---|
| PAM |   | 2 | 391 | 1065 | 1059 | 0.024 | -0.035 | 0.307 | 0.200 | 0.104 |
|---|
| ANP32E |   | 6 | 120 | 148 | 156 | -0.114 | 0.166 | -0.139 | 0.191 | 0.118 |
|---|
| DSG1 |   | 4 | 198 | 182 | 183 | -0.120 | 0.072 | -0.075 | 0.252 | 0.068 |
|---|
| PKIA |   | 3 | 265 | 832 | 816 | -0.091 | -0.120 | 0.085 | 0.249 | 0.044 |
|---|
GO Enrichment output for subnetwork 1100 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.857E-07 | 4.272E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.271E-07 | 3.762E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.271E-07 | 2.508E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 4.732E-07 | 2.721E-04 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 4.732E-07 | 2.177E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 5.831E-07 | 2.235E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.452E-07 | 2.12E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.527E-07 | 1.877E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 7.11E-07 | 1.817E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.585E-07 | 1.975E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.133E-06 | 2.369E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.866E-08 | 1.922E-04 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.767E-07 | 2.158E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 2.156E-07 | 1.755E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 2.605E-07 | 1.591E-04 |
|---|
| nuclear migration | GO:0007097 |  | 2.626E-07 | 1.283E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.626E-07 | 1.069E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.636E-07 | 9.199E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.12E-07 | 9.527E-05 |
|---|
| regulation of behavior | GO:0050795 |  | 3.707E-07 | 1.006E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.373E-07 | 1.068E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 4.587E-07 | 1.019E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.689E-08 | 1.85E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 1.742E-07 | 2.096E-04 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 1.742E-07 | 1.397E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 2.126E-07 | 1.278E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.577E-07 | 1.24E-04 |
|---|
| nuclear migration | GO:0007097 |  | 2.607E-07 | 1.045E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 2.607E-07 | 8.96E-05 |
|---|
| regulation of behavior | GO:0050795 |  | 3.076E-07 | 9.252E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.076E-07 | 8.224E-05 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.313E-07 | 1.038E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.554E-07 | 9.96E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.857E-07 | 4.272E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.271E-07 | 3.762E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.271E-07 | 2.508E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 4.732E-07 | 2.721E-04 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 4.732E-07 | 2.177E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 5.831E-07 | 2.235E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.452E-07 | 2.12E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.527E-07 | 1.877E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 7.11E-07 | 1.817E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.585E-07 | 1.975E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.133E-06 | 2.369E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.857E-07 | 4.272E-04 |
|---|
| nuclear migration | GO:0007097 |  | 3.271E-07 | 3.762E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 3.271E-07 | 2.508E-04 |
|---|
| positive regulation of locomotion | GO:0040017 |  | 4.732E-07 | 2.721E-04 |
|---|
| positive regulation of inflammatory response | GO:0050729 |  | 4.732E-07 | 2.177E-04 |
|---|
| regulation of chemotaxis | GO:0050920 |  | 5.831E-07 | 2.235E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.452E-07 | 2.12E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 6.527E-07 | 1.877E-04 |
|---|
| regulation of behavior | GO:0050795 |  | 7.11E-07 | 1.817E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 8.585E-07 | 1.975E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.133E-06 | 2.369E-04 |
|---|


