Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1091
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3359 | 1.341e-03 | 2.200e-05 | 1.543e-01 | 4.553e-09 |
|---|
| IPC | 0.3395 | 3.510e-01 | 1.804e-01 | 9.361e-01 | 5.927e-02 |
|---|
| Loi | 0.4701 | 7.400e-05 | 0.000e+00 | 5.668e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5934 | 1.000e-06 | 0.000e+00 | 1.269e-01 | 0.000e+00 |
|---|
| Wang | 0.2566 | 7.868e-03 | 6.753e-02 | 2.330e-01 | 1.238e-04 |
|---|
Expression data for subnetwork 1091 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
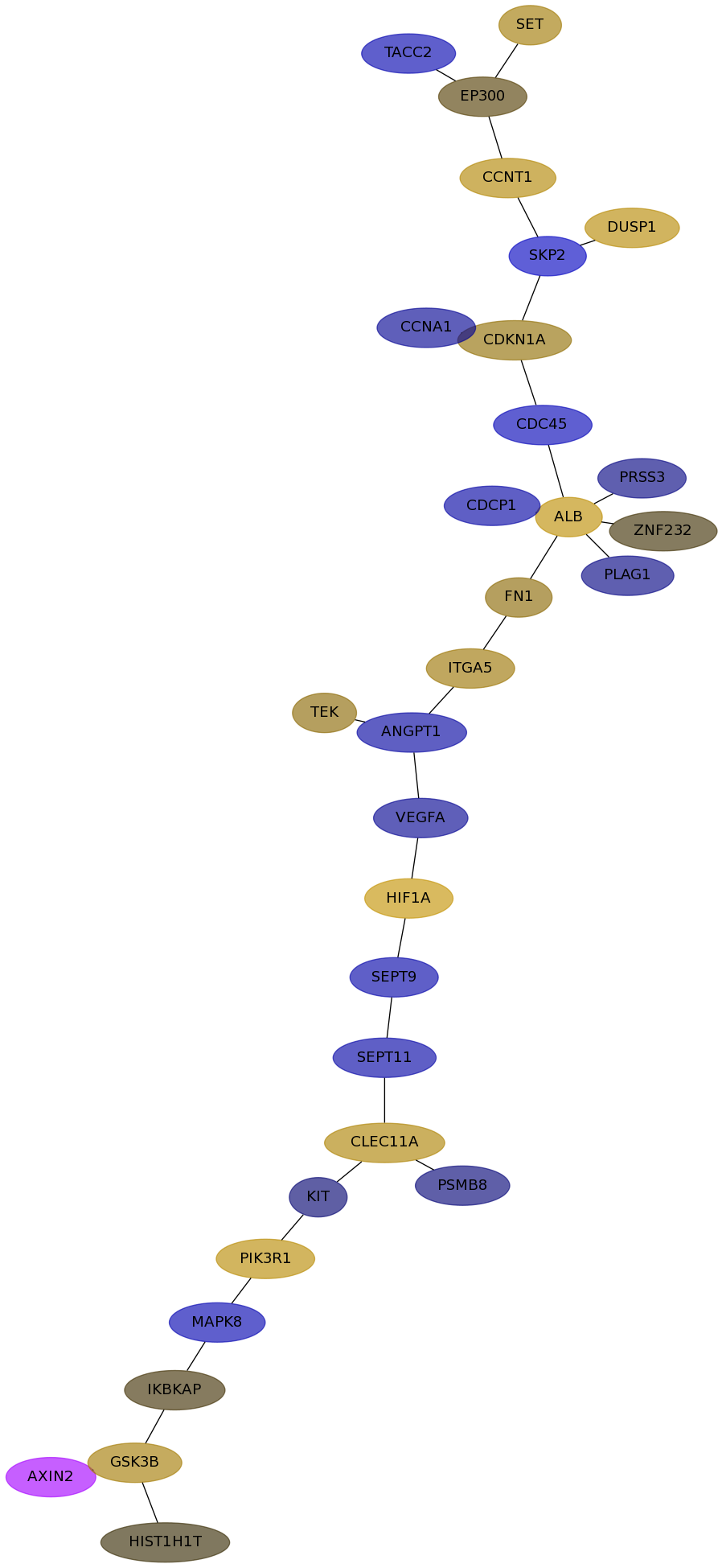
Score for each gene in subnetwork 1091 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| MAPK8 |   | 2 | 391 | 283 | 301 | -0.159 | 0.153 | 0.200 | 0.249 | 0.138 |
|---|
| TEK |   | 7 | 100 | 148 | 155 | 0.063 | -0.149 | -0.049 | 0.174 | 0.113 |
|---|
| CDCP1 |   | 2 | 391 | 57 | 80 | -0.117 | 0.068 | 0.045 | -0.058 | 0.010 |
|---|
| CLEC11A |   | 5 | 148 | 339 | 337 | 0.146 | 0.004 | 0.353 | 0.005 | 0.272 |
|---|
| FN1 |   | 5 | 148 | 57 | 63 | 0.064 | 0.190 | 0.133 | 0.091 | 0.083 |
|---|
| SEPT9 |   | 2 | 391 | 1063 | 1057 | -0.129 | -0.139 | -0.080 | 0.267 | -0.084 |
|---|
| PLAG1 |   | 1 | 652 | 1170 | 1177 | -0.053 | -0.092 | -0.040 | 0.304 | 0.021 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| PRSS3 |   | 1 | 652 | 1170 | 1177 | -0.052 | 0.151 | -0.165 | 0.007 | -0.001 |
|---|
| SKP2 |   | 14 | 42 | 148 | 146 | -0.228 | 0.092 | -0.043 | 0.237 | -0.009 |
|---|
| ITGA5 |   | 5 | 148 | 339 | 337 | 0.093 | 0.031 | 0.192 | 0.125 | 0.202 |
|---|
| CDKN1A |   | 56 | 4 | 1 | 3 | 0.075 | 0.064 | 0.254 | 0.030 | 0.082 |
|---|
| IKBKAP |   | 5 | 148 | 364 | 358 | 0.010 | 0.089 | 0.184 | 0.142 | -0.017 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| CDC45 |   | 4 | 198 | 847 | 830 | -0.177 | 0.134 | -0.171 | 0.185 | -0.085 |
|---|
| ZNF232 |   | 5 | 148 | 57 | 63 | 0.010 | 0.048 | -0.040 | 0.189 | 0.159 |
|---|
| PSMB8 |   | 4 | 198 | 710 | 696 | -0.039 | 0.254 | -0.071 | -0.088 | -0.139 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| ALB |   | 7 | 100 | 57 | 60 | 0.207 | -0.122 | 0.218 | 0.080 | 0.038 |
|---|
| HIST1H1T |   | 1 | 652 | 1170 | 1177 | 0.008 | 0.019 | 0.125 | -0.124 | -0.168 |
|---|
| TACC2 |   | 9 | 77 | 148 | 154 | -0.151 | -0.073 | 0.122 | 0.181 | 0.180 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| SET |   | 3 | 265 | 606 | 599 | 0.108 | 0.103 | 0.250 | 0.196 | -0.128 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| AXIN2 |   | 1 | 652 | 1170 | 1177 | undef | -0.151 | 0.143 | undef | undef |
|---|
| SEPT11 |   | 2 | 391 | 1063 | 1057 | -0.123 | -0.075 | -0.054 | 0.193 | 0.048 |
|---|
| GSK3B |   | 47 | 7 | 33 | 28 | 0.117 | -0.130 | 0.292 | -0.129 | 0.057 |
|---|
| KIT |   | 13 | 47 | 339 | 335 | -0.034 | -0.130 | -0.091 | 0.235 | -0.059 |
|---|
| ANGPT1 |   | 2 | 391 | 937 | 928 | -0.105 | 0.106 | -0.167 | 0.084 | 0.250 |
|---|
| DUSP1 |   | 1 | 652 | 1170 | 1177 | 0.178 | -0.019 | 0.127 | 0.170 | 0.134 |
|---|
GO Enrichment output for subnetwork 1091 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 8.107E-09 | 1.865E-05 |
|---|
| regulation of histone modification | GO:0031056 |  | 1.456E-08 | 1.674E-05 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 2.695E-07 | 2.066E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 7.25E-07 | 4.169E-04 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 8.178E-07 | 3.762E-04 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 1.404E-06 | 5.38E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 1.655E-06 | 5.437E-04 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.02E-06 | 5.808E-04 |
|---|
| zymogen activation | GO:0031638 |  | 4.309E-06 | 1.101E-03 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 5.91E-06 | 1.359E-03 |
|---|
| chromatin assembly | GO:0031497 |  | 7.295E-06 | 1.525E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 1.739E-09 | 4.249E-06 |
|---|
| regulation of histone modification | GO:0031056 |  | 3.125E-09 | 3.818E-06 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 1.454E-07 | 1.184E-04 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 3.601E-07 | 2.199E-04 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 4.22E-07 | 2.062E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 4.914E-07 | 2.001E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 6.332E-07 | 2.21E-04 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.352E-06 | 4.13E-04 |
|---|
| nucleosome assembly | GO:0006334 |  | 1.731E-06 | 4.698E-04 |
|---|
| chromatin assembly | GO:0031497 |  | 1.832E-06 | 4.475E-04 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 1.857E-06 | 4.123E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 2.086E-09 | 5.019E-06 |
|---|
| regulation of histone modification | GO:0031056 |  | 3.748E-09 | 4.509E-06 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 1.742E-07 | 1.397E-04 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 4.313E-07 | 2.594E-04 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 5.054E-07 | 2.432E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 5.885E-07 | 2.36E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 7.274E-07 | 2.5E-04 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 1.553E-06 | 4.672E-04 |
|---|
| nucleosome assembly | GO:0006334 |  | 1.917E-06 | 5.125E-04 |
|---|
| chromatin assembly | GO:0031497 |  | 2.032E-06 | 4.888E-04 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 2.132E-06 | 4.663E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 8.107E-09 | 1.865E-05 |
|---|
| regulation of histone modification | GO:0031056 |  | 1.456E-08 | 1.674E-05 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 2.695E-07 | 2.066E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 7.25E-07 | 4.169E-04 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 8.178E-07 | 3.762E-04 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 1.404E-06 | 5.38E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 1.655E-06 | 5.437E-04 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.02E-06 | 5.808E-04 |
|---|
| zymogen activation | GO:0031638 |  | 4.309E-06 | 1.101E-03 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 5.91E-06 | 1.359E-03 |
|---|
| chromatin assembly | GO:0031497 |  | 7.295E-06 | 1.525E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nucleosome disassembly | GO:0006337 |  | 8.107E-09 | 1.865E-05 |
|---|
| regulation of histone modification | GO:0031056 |  | 1.456E-08 | 1.674E-05 |
|---|
| cellular macromolecular complex disassembly | GO:0034623 |  | 2.695E-07 | 2.066E-04 |
|---|
| positive regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030949 |  | 7.25E-07 | 4.169E-04 |
|---|
| macromolecular complex disassembly | GO:0032984 |  | 8.178E-07 | 3.762E-04 |
|---|
| regulation of chromosome organization | GO:0033044 |  | 1.404E-06 | 5.38E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 1.655E-06 | 5.437E-04 |
|---|
| regulation of vascular endothelial growth factor receptor signaling pathway | GO:0030947 |  | 2.02E-06 | 5.808E-04 |
|---|
| zymogen activation | GO:0031638 |  | 4.309E-06 | 1.101E-03 |
|---|
| positive regulation of endothelial cell proliferation | GO:0001938 |  | 5.91E-06 | 1.359E-03 |
|---|
| chromatin assembly | GO:0031497 |  | 7.295E-06 | 1.525E-03 |
|---|


