Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1087
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3359 | 1.339e-03 | 2.200e-05 | 1.542e-01 | 4.543e-09 |
|---|
| IPC | 0.3406 | 3.491e-01 | 1.787e-01 | 9.357e-01 | 5.836e-02 |
|---|
| Loi | 0.4693 | 7.700e-05 | 0.000e+00 | 5.711e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5927 | 1.000e-06 | 0.000e+00 | 1.284e-01 | 0.000e+00 |
|---|
| Wang | 0.2569 | 7.757e-03 | 6.693e-02 | 2.315e-01 | 1.202e-04 |
|---|
Expression data for subnetwork 1087 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
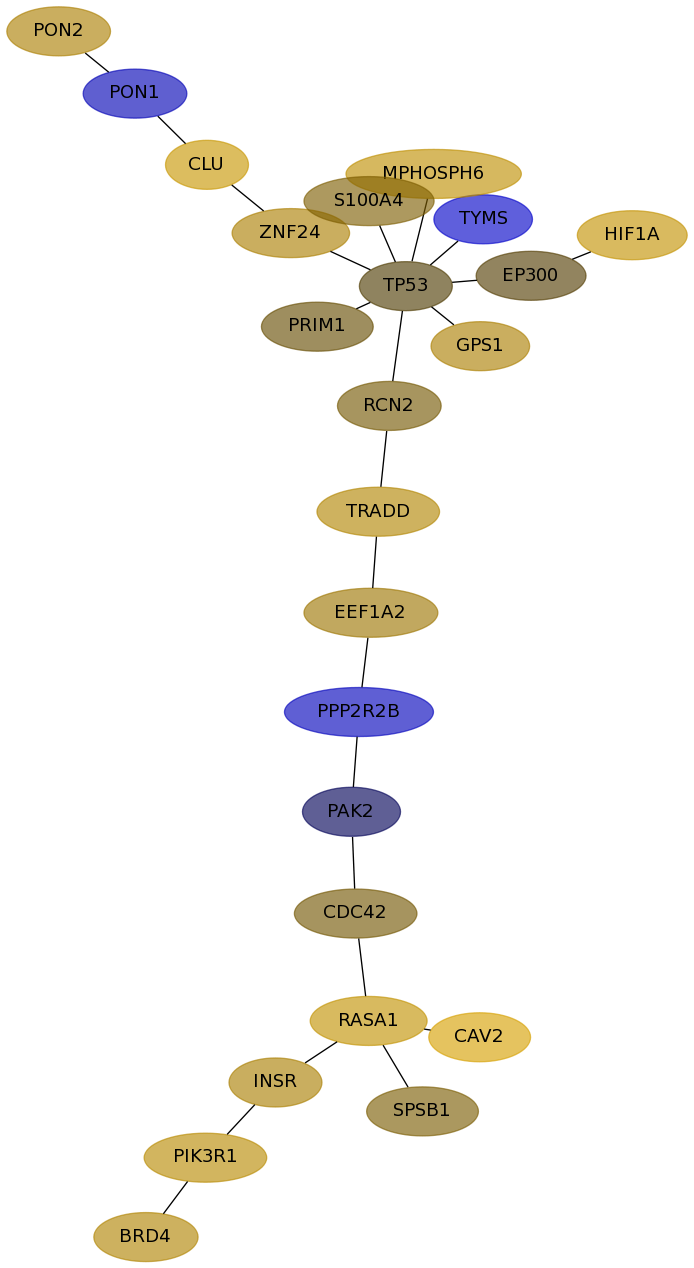
Score for each gene in subnetwork 1087 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| BRD4 |   | 18 | 26 | 1 | 12 | 0.156 | -0.119 | 0.095 | -0.075 | -0.044 |
|---|
| S100A4 |   | 6 | 120 | 283 | 282 | 0.047 | 0.163 | -0.116 | 0.096 | 0.148 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| SPSB1 |   | 7 | 100 | 492 | 495 | 0.044 | -0.124 | 0.088 | -0.152 | 0.169 |
|---|
| TRADD |   | 2 | 391 | 740 | 745 | 0.162 | 0.167 | 0.082 | -0.167 | 0.134 |
|---|
| RCN2 |   | 6 | 120 | 756 | 731 | 0.037 | 0.059 | 0.051 | 0.176 | -0.098 |
|---|
| PON1 |   | 1 | 652 | 1166 | 1169 | -0.172 | -0.182 | 0.160 | 0.086 | -0.114 |
|---|
| RASA1 |   | 9 | 77 | 492 | 494 | 0.237 | -0.079 | 0.146 | 0.127 | 0.149 |
|---|
| PRIM1 |   | 3 | 265 | 900 | 892 | 0.027 | 0.153 | -0.219 | 0.059 | 0.091 |
|---|
| PON2 |   | 1 | 652 | 1166 | 1169 | 0.138 | -0.263 | 0.185 | -0.141 | -0.048 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| ZNF24 |   | 1 | 652 | 1166 | 1169 | 0.137 | -0.198 | 0.080 | -0.018 | 0.162 |
|---|
| CDC42 |   | 44 | 9 | 1 | 6 | 0.036 | 0.047 | 0.242 | -0.027 | -0.042 |
|---|
| HIF1A |   | 21 | 23 | 33 | 31 | 0.246 | -0.098 | 0.224 | -0.118 | 0.356 |
|---|
| EEF1A2 |   | 9 | 77 | 182 | 179 | 0.098 | 0.032 | 0.220 | -0.119 | 0.102 |
|---|
| PAK2 |   | 20 | 24 | 1 | 11 | -0.019 | 0.014 | 0.161 | 0.081 | 0.062 |
|---|
| TYMS |   | 1 | 652 | 1166 | 1169 | -0.273 | 0.119 | -0.217 | 0.090 | 0.014 |
|---|
| PPP2R2B |   | 35 | 13 | 1 | 8 | -0.200 | 0.124 | -0.019 | 0.029 | -0.073 |
|---|
| CLU |   | 10 | 67 | 117 | 121 | 0.280 | -0.208 | 0.119 | -0.153 | -0.234 |
|---|
| GPS1 |   | 3 | 265 | 900 | 892 | 0.137 | -0.239 | -0.126 | 0.057 | -0.145 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| MPHOSPH6 |   | 8 | 84 | 756 | 726 | 0.217 | -0.084 | 0.175 | -0.269 | 0.146 |
|---|
GO Enrichment output for subnetwork 1087 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.699E-07 | 3.907E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.699E-07 | 1.954E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.699E-07 | 1.302E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 2.896E-07 | 1.665E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.391E-07 | 1.56E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 3.391E-07 | 1.3E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.924E-07 | 1.946E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.924E-07 | 1.703E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 1.416E-06 | 3.62E-04 |
|---|
| regulation of glycolysis | GO:0006110 |  | 1.416E-06 | 3.258E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.416E-06 | 2.962E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 6.678E-08 | 1.632E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.334E-07 | 1.629E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.334E-07 | 1.086E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.423E-07 | 8.69E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.725E-07 | 1.82E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 3.725E-07 | 1.517E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 3.725E-07 | 1.3E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 5.579E-07 | 1.704E-04 |
|---|
| regulation of glycolysis | GO:0006110 |  | 5.579E-07 | 1.514E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 5.579E-07 | 1.363E-04 |
|---|
| regulation of cellular carbohydrate catabolic process | GO:0043471 |  | 7.959E-07 | 1.768E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 7.496E-08 | 1.803E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 1.445E-07 | 1.739E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.497E-07 | 1.201E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.497E-07 | 9.004E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.616E-07 | 1.259E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.616E-07 | 1.049E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 4.18E-07 | 1.437E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 6.261E-07 | 1.883E-04 |
|---|
| regulation of glycolysis | GO:0006110 |  | 6.261E-07 | 1.674E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 6.261E-07 | 1.506E-04 |
|---|
| regulation of cellular carbohydrate catabolic process | GO:0043471 |  | 8.931E-07 | 1.953E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.699E-07 | 3.907E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.699E-07 | 1.954E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.699E-07 | 1.302E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 2.896E-07 | 1.665E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.391E-07 | 1.56E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 3.391E-07 | 1.3E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.924E-07 | 1.946E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.924E-07 | 1.703E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 1.416E-06 | 3.62E-04 |
|---|
| regulation of glycolysis | GO:0006110 |  | 1.416E-06 | 3.258E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.416E-06 | 2.962E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of glycolysis | GO:0045821 |  | 1.699E-07 | 3.907E-04 |
|---|
| nuclear migration | GO:0007097 |  | 1.699E-07 | 1.954E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.699E-07 | 1.302E-04 |
|---|
| regulation of protein complex assembly | GO:0043254 |  | 2.896E-07 | 1.665E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 3.391E-07 | 1.56E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 3.391E-07 | 1.3E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 5.924E-07 | 1.946E-04 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 5.924E-07 | 1.703E-04 |
|---|
| positive regulation of glucose metabolic process | GO:0010907 |  | 1.416E-06 | 3.62E-04 |
|---|
| regulation of glycolysis | GO:0006110 |  | 1.416E-06 | 3.258E-04 |
|---|
| nucleus localization | GO:0051647 |  | 1.416E-06 | 2.962E-04 |
|---|


