Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1021
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3408 | 1.073e-03 | 1.500e-05 | 1.400e-01 | 2.253e-09 |
|---|
| IPC | 0.3385 | 3.527e-01 | 1.818e-01 | 9.365e-01 | 6.006e-02 |
|---|
| Loi | 0.4606 | 1.130e-04 | 0.000e+00 | 6.182e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5831 | 2.000e-06 | 0.000e+00 | 1.511e-01 | 0.000e+00 |
|---|
| Wang | 0.2538 | 8.809e-03 | 7.246e-02 | 2.454e-01 | 1.566e-04 |
|---|
Expression data for subnetwork 1021 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
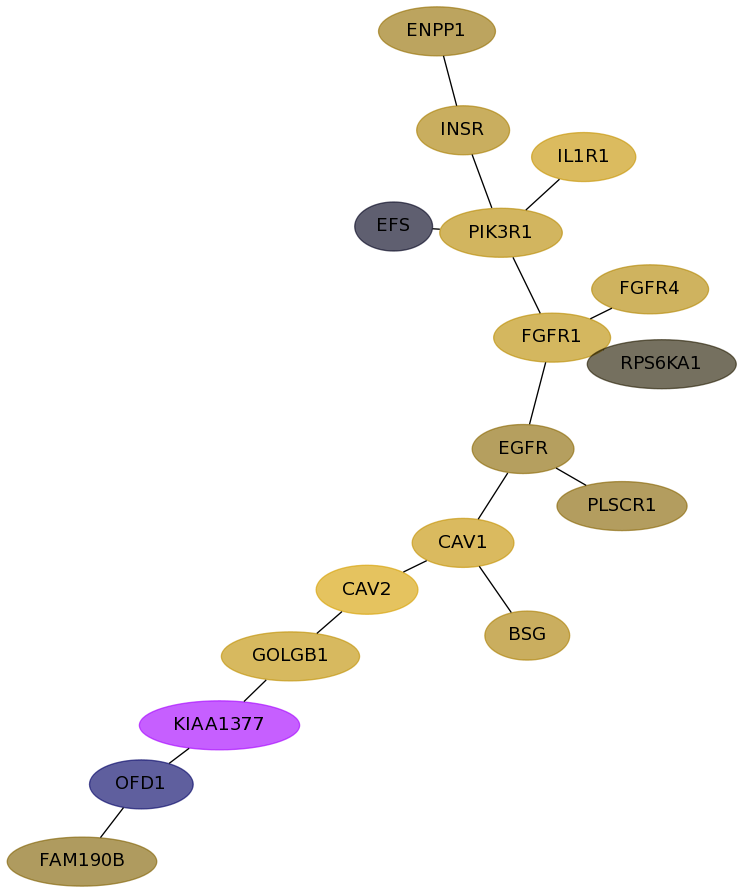
Score for each gene in subnetwork 1021 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| IL1R1 |   | 8 | 84 | 536 | 515 | 0.257 | -0.211 | 0.178 | -0.071 | 0.003 |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| ENPP1 |   | 3 | 265 | 778 | 770 | 0.082 | 0.088 | 0.129 | -0.026 | 0.039 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| PLSCR1 |   | 7 | 100 | 634 | 614 | 0.058 | 0.202 | -0.159 | 0.091 | -0.105 |
|---|
| RPS6KA1 |   | 8 | 84 | 696 | 669 | 0.005 | -0.208 | 0.084 | -0.322 | -0.071 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| OFD1 |   | 1 | 652 | 1411 | 1411 | -0.027 | -0.056 | 0.206 | -0.135 | -0.077 |
|---|
| BSG |   | 1 | 652 | 1411 | 1411 | 0.138 | -0.039 | 0.061 | -0.119 | -0.040 |
|---|
| EFS |   | 4 | 198 | 129 | 130 | -0.003 | -0.024 | 0.300 | -0.003 | -0.034 |
|---|
| CAV1 |   | 10 | 67 | 439 | 428 | 0.251 | -0.092 | 0.124 | 0.079 | 0.119 |
|---|
| KIAA1377 |   | 1 | 652 | 1411 | 1411 | undef | -0.095 | 0.116 | undef | undef |
|---|
| FAM190B |   | 1 | 652 | 1411 | 1411 | 0.051 | -0.032 | 0.110 | 0.180 | 0.140 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| GOLGB1 |   | 3 | 265 | 117 | 127 | 0.231 | 0.104 | 0.352 | 0.044 | 0.175 |
|---|
GO Enrichment output for subnetwork 1021 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.622E-10 | 8.331E-07 |
|---|
| lipid localization | GO:0010876 |  | 9.501E-09 | 1.093E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 7.331E-08 | 5.621E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 7.331E-08 | 4.215E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 1.464E-07 | 6.735E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 1.464E-07 | 5.613E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 1.464E-07 | 4.811E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.559E-07 | 7.357E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 5.981E-07 | 1.529E-04 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 6.124E-07 | 1.409E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 6.124E-07 | 1.281E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.768E-10 | 4.319E-07 |
|---|
| lipid localization | GO:0010876 |  | 5.876E-09 | 7.177E-06 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 3.901E-08 | 3.177E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 1.362E-07 | 8.32E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 1.362E-07 | 6.656E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 2.177E-07 | 8.864E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 2.177E-07 | 7.598E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 2.177E-07 | 6.648E-05 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 3.262E-07 | 8.854E-05 |
|---|
| tissue remodeling | GO:0048771 |  | 3.772E-07 | 9.215E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 4.211E-07 | 9.352E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.731E-10 | 4.165E-07 |
|---|
| lipid localization | GO:0010876 |  | 5.155E-09 | 6.202E-06 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 4.564E-08 | 3.66E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 9.116E-08 | 5.483E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 1.593E-07 | 7.668E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 1.593E-07 | 6.39E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 1.593E-07 | 5.477E-05 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 3.815E-07 | 1.147E-04 |
|---|
| gas homeostasis | GO:0033483 |  | 3.815E-07 | 1.02E-04 |
|---|
| tissue remodeling | GO:0048771 |  | 4.621E-07 | 1.112E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 4.621E-07 | 1.011E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.622E-10 | 8.331E-07 |
|---|
| lipid localization | GO:0010876 |  | 9.501E-09 | 1.093E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 7.331E-08 | 5.621E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 7.331E-08 | 4.215E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 1.464E-07 | 6.735E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 1.464E-07 | 5.613E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 1.464E-07 | 4.811E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.559E-07 | 7.357E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 5.981E-07 | 1.529E-04 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 6.124E-07 | 1.409E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 6.124E-07 | 1.281E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 3.622E-10 | 8.331E-07 |
|---|
| lipid localization | GO:0010876 |  | 9.501E-09 | 1.093E-05 |
|---|
| positive regulation of vasoconstriction | GO:0045907 |  | 7.331E-08 | 5.621E-05 |
|---|
| positive regulation of microtubule polymerization | GO:0031116 |  | 7.331E-08 | 4.215E-05 |
|---|
| gas homeostasis | GO:0033483 |  | 1.464E-07 | 6.735E-05 |
|---|
| positive regulation of microtubule polymerization or depolymerization | GO:0031112 |  | 1.464E-07 | 5.613E-05 |
|---|
| regulation of microtubule polymerization | GO:0031113 |  | 1.464E-07 | 4.811E-05 |
|---|
| negative regulation of JAK-STAT cascade | GO:0046426 |  | 2.559E-07 | 7.357E-05 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 5.981E-07 | 1.529E-04 |
|---|
| regulation of epithelial cell differentiation | GO:0030856 |  | 6.124E-07 | 1.409E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 6.124E-07 | 1.281E-04 |
|---|


