Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1017
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3409 | 1.066e-03 | 1.500e-05 | 1.396e-01 | 2.233e-09 |
|---|
| IPC | 0.3393 | 3.513e-01 | 1.806e-01 | 9.362e-01 | 5.942e-02 |
|---|
| Loi | 0.4599 | 1.170e-04 | 0.000e+00 | 6.223e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5833 | 2.000e-06 | 0.000e+00 | 1.506e-01 | 0.000e+00 |
|---|
| Wang | 0.2536 | 8.872e-03 | 7.278e-02 | 2.462e-01 | 1.590e-04 |
|---|
Expression data for subnetwork 1017 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
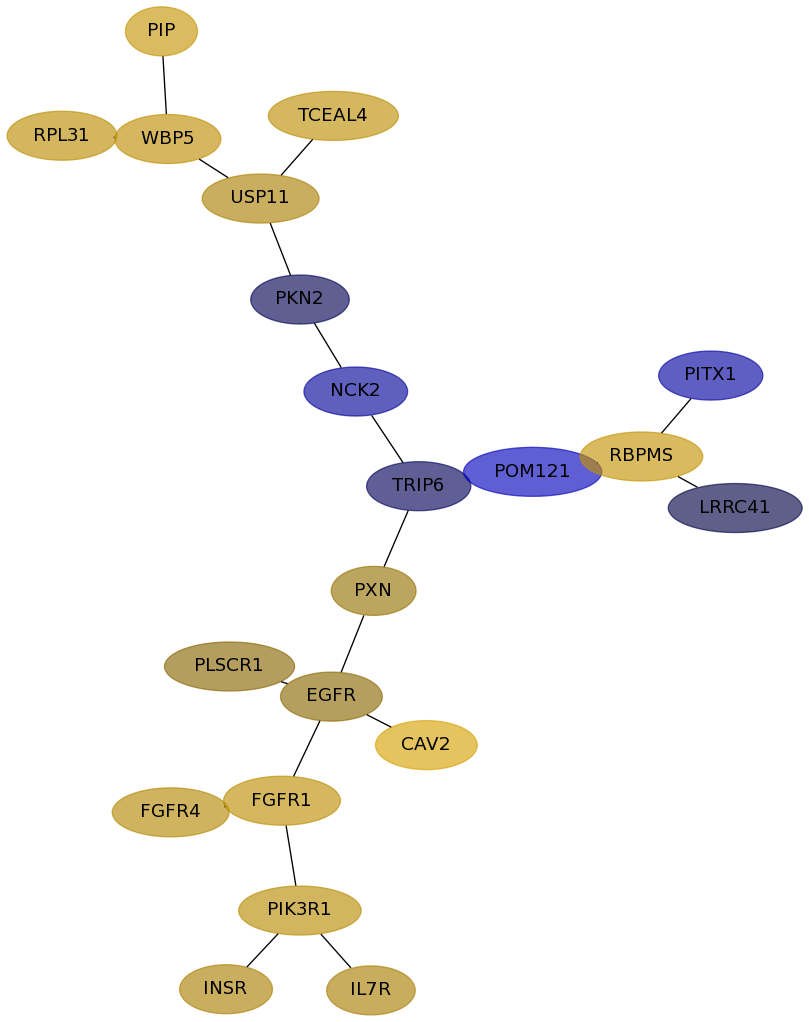
Score for each gene in subnetwork 1017 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FGFR4 |   | 37 | 12 | 262 | 262 | 0.171 | -0.011 | 0.188 | -0.089 | -0.049 |
|---|
| PXN |   | 5 | 148 | 439 | 432 | 0.085 | -0.088 | 0.284 | -0.022 | 0.114 |
|---|
| PLSCR1 |   | 7 | 100 | 634 | 614 | 0.058 | 0.202 | -0.159 | 0.091 | -0.105 |
|---|
| TRIP6 |   | 3 | 265 | 364 | 362 | -0.020 | 0.170 | 0.145 | 0.034 | -0.213 |
|---|
| RPL31 |   | 2 | 391 | 823 | 817 | 0.200 | -0.027 | -0.118 | 0.113 | 0.003 |
|---|
| PIK3R1 |   | 91 | 1 | 1 | 1 | 0.189 | -0.237 | 0.381 | 0.065 | -0.002 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FGFR1 |   | 41 | 10 | 129 | 123 | 0.205 | -0.036 | 0.200 | 0.087 | -0.015 |
|---|
| PIP |   | 2 | 391 | 823 | 817 | 0.253 | -0.166 | 0.192 | -0.153 | -0.117 |
|---|
| POM121 |   | 5 | 148 | 419 | 415 | -0.195 | -0.197 | 0.101 | 0.079 | 0.017 |
|---|
| PKN2 |   | 3 | 265 | 552 | 550 | -0.017 | -0.198 | 0.158 | 0.060 | 0.068 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| IL7R |   | 14 | 42 | 339 | 334 | 0.130 | -0.151 | -0.112 | -0.130 | -0.160 |
|---|
| PITX1 |   | 2 | 391 | 1410 | 1391 | -0.105 | 0.256 | 0.046 | 0.091 | 0.139 |
|---|
| TCEAL4 |   | 3 | 265 | 552 | 550 | 0.205 | -0.156 | 0.219 | 0.091 | 0.007 |
|---|
| NCK2 |   | 2 | 391 | 262 | 283 | -0.089 | -0.091 | 0.064 | 0.021 | -0.053 |
|---|
| USP11 |   | 5 | 148 | 283 | 287 | 0.134 | -0.144 | 0.106 | 0.121 | -0.044 |
|---|
| RBPMS |   | 7 | 100 | 419 | 407 | 0.246 | 0.043 | 0.108 | 0.066 | -0.077 |
|---|
| CAV2 |   | 52 | 6 | 117 | 118 | 0.380 | -0.013 | 0.165 | 0.065 | 0.101 |
|---|
| LRRC41 |   | 3 | 265 | 419 | 419 | -0.012 | -0.136 | 0.210 | -0.080 | -0.125 |
|---|
| WBP5 |   | 2 | 391 | 823 | 817 | 0.214 | -0.089 | 0.073 | 0.201 | 0.142 |
|---|
GO Enrichment output for subnetwork 1017 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.642E-07 | 3.776E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.236E-06 | 1.422E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.18E-06 | 3.205E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 8.001E-06 | 4.6E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 9.869E-06 | 4.54E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 9.869E-06 | 3.783E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.165E-05 | 3.828E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.182E-05 | 3.399E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.902E-05 | 7.415E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.902E-05 | 6.674E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 3.714E-05 | 7.766E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.911E-08 | 1.2E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.815E-06 | 2.216E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.317E-06 | 2.701E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 3.377E-06 | 2.063E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.048E-06 | 1.978E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.048E-06 | 1.648E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 5.245E-06 | 1.831E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 9.965E-06 | 3.043E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.435E-05 | 3.894E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.605E-05 | 3.921E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.788E-05 | 3.971E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 5.079E-08 | 1.222E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.719E-06 | 2.068E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 3.934E-06 | 3.155E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.065E-06 | 2.445E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 4.065E-06 | 1.956E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 4.065E-06 | 1.63E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 6.268E-06 | 2.154E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 9.145E-06 | 2.75E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.36E-05 | 3.637E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.536E-05 | 3.695E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 1.726E-05 | 3.774E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.642E-07 | 3.776E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.236E-06 | 1.422E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.18E-06 | 3.205E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 8.001E-06 | 4.6E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 9.869E-06 | 4.54E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 9.869E-06 | 3.783E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.165E-05 | 3.828E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.182E-05 | 3.399E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.902E-05 | 7.415E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.902E-05 | 6.674E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 3.714E-05 | 7.766E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.642E-07 | 3.776E-04 |
|---|
| positive regulation of protein polymerization | GO:0032273 |  | 1.236E-06 | 1.422E-03 |
|---|
| positive regulation of protein complex assembly | GO:0031334 |  | 4.18E-06 | 3.205E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 8.001E-06 | 4.6E-03 |
|---|
| signal complex assembly | GO:0007172 |  | 9.869E-06 | 4.54E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 9.869E-06 | 3.783E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.165E-05 | 3.828E-03 |
|---|
| positive regulation of cytoskeleton organization | GO:0051495 |  | 1.182E-05 | 3.399E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.902E-05 | 7.415E-03 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.902E-05 | 6.674E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 3.714E-05 | 7.766E-03 |
|---|


