Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1013
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3402 | 1.100e-03 | 1.600e-05 | 1.415e-01 | 2.491e-09 |
|---|
| IPC | 0.3404 | 3.495e-01 | 1.790e-01 | 9.358e-01 | 5.854e-02 |
|---|
| Loi | 0.4598 | 1.170e-04 | 0.000e+00 | 6.226e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5824 | 2.000e-06 | 0.000e+00 | 1.528e-01 | 0.000e+00 |
|---|
| Wang | 0.2541 | 8.698e-03 | 7.188e-02 | 2.440e-01 | 1.525e-04 |
|---|
Expression data for subnetwork 1013 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
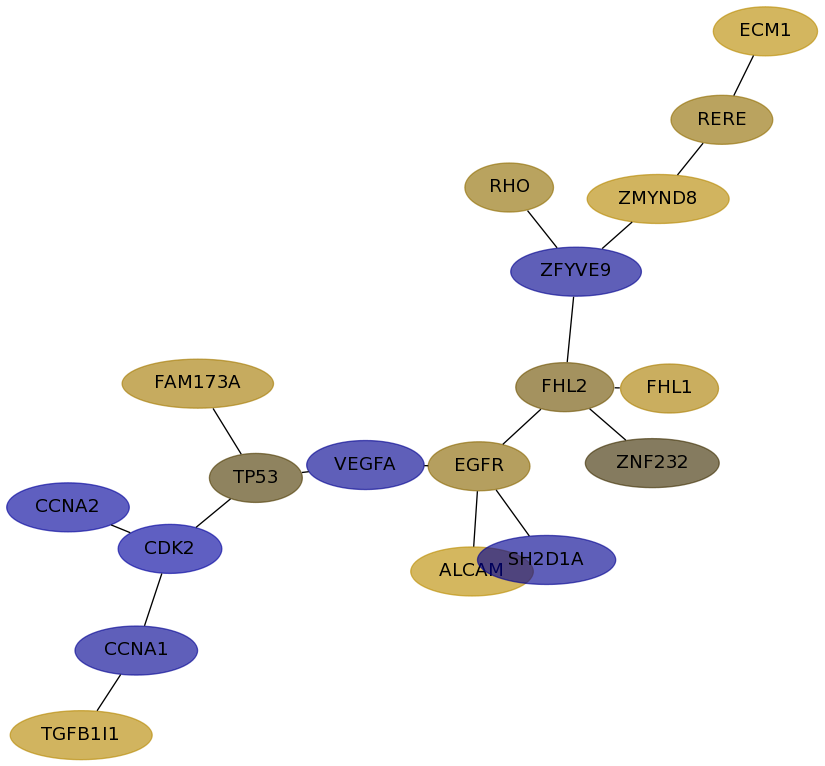
Score for each gene in subnetwork 1013 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| FHL2 |   | 5 | 148 | 206 | 202 | 0.033 | 0.143 | 0.239 | -0.033 | 0.294 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| ALCAM |   | 4 | 198 | 1276 | 1276 | 0.206 | -0.208 | 0.193 | 0.067 | -0.010 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| ZFYVE9 |   | 1 | 652 | 1406 | 1407 | -0.070 | -0.247 | 0.042 | -0.003 | 0.054 |
|---|
| RERE |   | 4 | 198 | 634 | 615 | 0.077 | -0.284 | 0.209 | -0.027 | 0.074 |
|---|
| FAM173A |   | 3 | 265 | 1325 | 1319 | 0.118 | 0.125 | -0.114 | -0.218 | 0.169 |
|---|
| ZNF232 |   | 5 | 148 | 57 | 63 | 0.010 | 0.048 | -0.040 | 0.189 | 0.159 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| ECM1 |   | 1 | 652 | 1406 | 1407 | 0.197 | 0.107 | 0.148 | -0.144 | 0.048 |
|---|
| SH2D1A |   | 2 | 391 | 584 | 578 | -0.074 | -0.078 | -0.261 | -0.082 | -0.033 |
|---|
| CCNA1 |   | 17 | 30 | 326 | 314 | -0.078 | 0.174 | 0.092 | 0.078 | -0.098 |
|---|
| RHO |   | 1 | 652 | 1406 | 1407 | 0.075 | -0.033 | -0.036 | -0.118 | -0.081 |
|---|
| ZMYND8 |   | 1 | 652 | 1406 | 1407 | 0.178 | -0.140 | 0.259 | -0.112 | 0.011 |
|---|
| FHL1 |   | 3 | 265 | 1292 | 1278 | 0.136 | -0.041 | 0.104 | 0.301 | 0.266 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
| TGFB1I1 |   | 16 | 34 | 326 | 315 | 0.176 | -0.105 | 0.229 | 0.143 | 0.298 |
|---|
GO Enrichment output for subnetwork 1013 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 2.216E-05 | 0.05095657 |
|---|
| protein import into nucleus | GO:0006606 |  | 3.532E-05 | 0.0406179 |
|---|
| nuclear import | GO:0051170 |  | 3.724E-05 | 0.0285519 |
|---|
| protein localization in nucleus | GO:0034504 |  | 4.802E-05 | 0.02761085 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 6.161E-05 | 0.02834218 |
|---|
| SMAD protein complex assembly | GO:0007183 |  | 6.161E-05 | 0.02361848 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 6.161E-05 | 0.02024441 |
|---|
| phototransduction. visible light | GO:0007603 |  | 6.161E-05 | 0.01771386 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 6.161E-05 | 0.01574565 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.161E-05 | 0.01417109 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 6.365E-05 | 0.013309 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 1.077E-05 | 0.02630439 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.242E-05 | 0.01517145 |
|---|
| nuclear import | GO:0051170 |  | 1.303E-05 | 0.01061281 |
|---|
| protein localization in nucleus | GO:0034504 |  | 1.718E-05 | 0.0104949 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 2.887E-05 | 0.0141082 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 2.887E-05 | 0.01175684 |
|---|
| regulation of phospholipase A2 activity | GO:0032429 |  | 2.887E-05 | 0.01007729 |
|---|
| G2/M transition checkpoint | GO:0031576 |  | 2.887E-05 | 8.818E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 2.887E-05 | 7.838E-03 |
|---|
| protein insertion into membrane | GO:0051205 |  | 2.887E-05 | 7.054E-03 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.027E-05 | 6.722E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 1.199E-05 | 0.02884163 |
|---|
| protein import into nucleus | GO:0006606 |  | 1.526E-05 | 0.01835864 |
|---|
| nuclear import | GO:0051170 |  | 1.603E-05 | 0.01285715 |
|---|
| protein localization in nucleus | GO:0034504 |  | 2.127E-05 | 0.01279475 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 3.193E-05 | 0.01536593 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 3.384E-05 | 0.01356838 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 3.384E-05 | 0.01163004 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 3.384E-05 | 0.01017628 |
|---|
| regulation of phospholipase A2 activity | GO:0032429 |  | 3.384E-05 | 9.046E-03 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 3.384E-05 | 8.141E-03 |
|---|
| DNA integrity checkpoint | GO:0031570 |  | 4.162E-05 | 9.104E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 2.216E-05 | 0.05095657 |
|---|
| protein import into nucleus | GO:0006606 |  | 3.532E-05 | 0.0406179 |
|---|
| nuclear import | GO:0051170 |  | 3.724E-05 | 0.0285519 |
|---|
| protein localization in nucleus | GO:0034504 |  | 4.802E-05 | 0.02761085 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 6.161E-05 | 0.02834218 |
|---|
| SMAD protein complex assembly | GO:0007183 |  | 6.161E-05 | 0.02361848 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 6.161E-05 | 0.02024441 |
|---|
| phototransduction. visible light | GO:0007603 |  | 6.161E-05 | 0.01771386 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 6.161E-05 | 0.01574565 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.161E-05 | 0.01417109 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 6.365E-05 | 0.013309 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 2.216E-05 | 0.05095657 |
|---|
| protein import into nucleus | GO:0006606 |  | 3.532E-05 | 0.0406179 |
|---|
| nuclear import | GO:0051170 |  | 3.724E-05 | 0.0285519 |
|---|
| protein localization in nucleus | GO:0034504 |  | 4.802E-05 | 0.02761085 |
|---|
| negative regulation of bone resorption | GO:0045779 |  | 6.161E-05 | 0.02834218 |
|---|
| SMAD protein complex assembly | GO:0007183 |  | 6.161E-05 | 0.02361848 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 6.161E-05 | 0.02024441 |
|---|
| phototransduction. visible light | GO:0007603 |  | 6.161E-05 | 0.01771386 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 6.161E-05 | 0.01574565 |
|---|
| positive regulation of blood vessel endothelial cell migration | GO:0043536 |  | 6.161E-05 | 0.01417109 |
|---|
| DNA damage checkpoint | GO:0000077 |  | 6.365E-05 | 0.013309 |
|---|


