Study VanDeVijver-ER-neg
Study informations
122 subnetworks in total page | file
1481 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1001
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| Desmedt | 0.3381 | 1.215e-03 | 1.800e-05 | 1.478e-01 | 3.233e-09 |
|---|
| IPC | 0.3479 | 3.371e-01 | 1.683e-01 | 9.332e-01 | 5.296e-02 |
|---|
| Loi | 0.4578 | 1.280e-04 | 0.000e+00 | 6.340e-02 | 0.000e+00 |
|---|
| Schmidt | 0.5859 | 2.000e-06 | 0.000e+00 | 1.444e-01 | 0.000e+00 |
|---|
| Wang | 0.2544 | 8.591e-03 | 7.133e-02 | 2.426e-01 | 1.487e-04 |
|---|
Expression data for subnetwork 1001 in each dataset
Desmedt |
IPC |
Loi |
Schmidt |
Wang |
Subnetwork structure for each dataset
- Desmedt
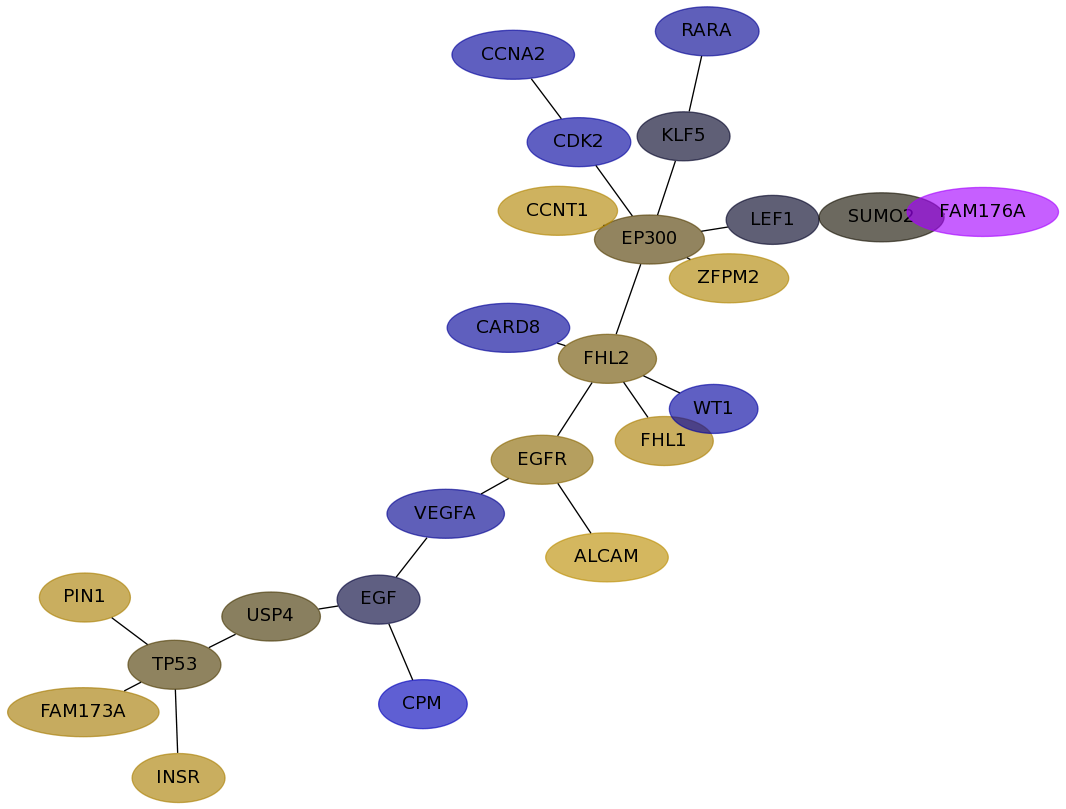
Score for each gene in subnetwork 1001 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC | Loi | Schmidt | Wang |
|---|
| EP300 |   | 71 | 3 | 1 | 2 | 0.016 | -0.149 | 0.087 | 0.102 | 0.025 |
|---|
| USP4 |   | 6 | 120 | 823 | 803 | 0.012 | -0.093 | 0.147 | 0.097 | 0.054 |
|---|
| FAM176A |   | 2 | 391 | 1065 | 1059 | undef | 0.054 | 0.217 | undef | undef |
|---|
| RARA |   | 1 | 652 | 1325 | 1328 | -0.076 | 0.024 | -0.028 | -0.056 | 0.110 |
|---|
| SUMO2 |   | 12 | 55 | 148 | 150 | 0.003 | 0.117 | 0.062 | 0.294 | -0.096 |
|---|
| EGF |   | 11 | 59 | 398 | 383 | -0.009 | -0.212 | 0.182 | -0.276 | 0.005 |
|---|
| FHL2 |   | 5 | 148 | 206 | 202 | 0.033 | 0.143 | 0.239 | -0.033 | 0.294 |
|---|
| TP53 |   | 28 | 15 | 1 | 9 | 0.015 | -0.167 | 0.148 | -0.027 | 0.147 |
|---|
| ZFPM2 |   | 4 | 198 | 148 | 162 | 0.160 | 0.001 | 0.092 | 0.113 | 0.379 |
|---|
| VEGFA |   | 28 | 15 | 57 | 49 | -0.074 | 0.090 | 0.200 | 0.030 | 0.148 |
|---|
| ALCAM |   | 4 | 198 | 1276 | 1276 | 0.206 | -0.208 | 0.193 | 0.067 | -0.010 |
|---|
| EGFR |   | 72 | 2 | 117 | 117 | 0.064 | -0.195 | 0.240 | -0.205 | 0.065 |
|---|
| FAM173A |   | 3 | 265 | 1325 | 1319 | 0.118 | 0.125 | -0.114 | -0.218 | 0.169 |
|---|
| INSR |   | 47 | 7 | 1 | 5 | 0.135 | 0.063 | 0.173 | 0.135 | -0.072 |
|---|
| CCNA2 |   | 15 | 40 | 148 | 144 | -0.097 | 0.176 | -0.053 | 0.188 | 0.246 |
|---|
| LEF1 |   | 2 | 391 | 1325 | 1321 | -0.005 | -0.068 | 0.016 | -0.018 | 0.139 |
|---|
| CARD8 |   | 1 | 652 | 1325 | 1328 | -0.089 | -0.143 | -0.168 | -0.081 | 0.113 |
|---|
| CCNT1 |   | 18 | 26 | 33 | 32 | 0.163 | 0.008 | -0.028 | 0.162 | -0.213 |
|---|
| PIN1 |   | 17 | 30 | 1 | 13 | 0.135 | 0.194 | 0.040 | -0.040 | -0.048 |
|---|
| CPM |   | 2 | 391 | 620 | 618 | -0.196 | 0.166 | -0.062 | -0.079 | 0.280 |
|---|
| WT1 |   | 8 | 84 | 283 | 278 | -0.106 | 0.254 | -0.039 | 0.036 | 0.172 |
|---|
| KLF5 |   | 25 | 19 | 57 | 50 | -0.005 | 0.077 | 0.117 | -0.007 | -0.006 |
|---|
| FHL1 |   | 3 | 265 | 1292 | 1278 | 0.136 | -0.041 | 0.104 | 0.301 | 0.266 |
|---|
| CDK2 |   | 16 | 34 | 129 | 124 | -0.104 | 0.196 | -0.096 | 0.109 | 0.362 |
|---|
GO Enrichment output for subnetwork 1001 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 2.767E-06 | 6.363E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.137E-05 | 0.01307505 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.451E-05 | 0.01112294 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.545E-05 | 8.886E-03 |
|---|
| somitogenesis | GO:0001756 |  | 1.851E-05 | 8.513E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.11E-05 | 8.087E-03 |
|---|
| sex determination | GO:0007530 |  | 2.193E-05 | 7.206E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.575E-05 | 7.402E-03 |
|---|
| vasculogenesis | GO:0001570 |  | 3.464E-05 | 8.852E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 3.464E-05 | 7.967E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 5.8E-05 | 0.01212754 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of I-kappaB kinase/NF-kappaB cascade | GO:0043124 |  | 6.36E-07 | 1.554E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 9.073E-07 | 1.108E-03 |
|---|
| positive regulation of interleukin-1 beta secretion | GO:0050718 |  | 9.073E-07 | 7.389E-04 |
|---|
| positive regulation of interleukin-1 secretion | GO:0050716 |  | 1.246E-06 | 7.609E-04 |
|---|
| regulation of interleukin-1 beta secretion | GO:0050706 |  | 1.246E-06 | 6.087E-04 |
|---|
| regulation of protein secretion | GO:0050708 |  | 1.544E-06 | 6.289E-04 |
|---|
| regulation of interleukin-1 secretion | GO:0050704 |  | 1.659E-06 | 5.789E-04 |
|---|
| positive regulation of cytokine secretion | GO:0050715 |  | 4.198E-06 | 1.282E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.962E-06 | 1.347E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.329E-06 | 1.302E-03 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.715E-06 | 1.269E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of I-kappaB kinase/NF-kappaB cascade | GO:0043124 |  | 8.755E-07 | 2.106E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 1.249E-06 | 1.502E-03 |
|---|
| positive regulation of interleukin-1 beta secretion | GO:0050718 |  | 1.249E-06 | 1.001E-03 |
|---|
| positive regulation of interleukin-1 secretion | GO:0050716 |  | 1.714E-06 | 1.031E-03 |
|---|
| regulation of interleukin-1 beta secretion | GO:0050706 |  | 1.714E-06 | 8.249E-04 |
|---|
| regulation of interleukin-1 secretion | GO:0050704 |  | 2.282E-06 | 9.151E-04 |
|---|
| regulation of protein secretion | GO:0050708 |  | 2.349E-06 | 8.074E-04 |
|---|
| positive regulation of cytokine secretion | GO:0050715 |  | 5.772E-06 | 1.736E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 6.998E-06 | 1.871E-03 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 7.003E-06 | 1.685E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 7.53E-06 | 1.647E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 2.767E-06 | 6.363E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.137E-05 | 0.01307505 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.451E-05 | 0.01112294 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.545E-05 | 8.886E-03 |
|---|
| somitogenesis | GO:0001756 |  | 1.851E-05 | 8.513E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.11E-05 | 8.087E-03 |
|---|
| sex determination | GO:0007530 |  | 2.193E-05 | 7.206E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.575E-05 | 7.402E-03 |
|---|
| vasculogenesis | GO:0001570 |  | 3.464E-05 | 8.852E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 3.464E-05 | 7.967E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 5.8E-05 | 0.01212754 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein sumoylation | GO:0016925 |  | 2.767E-06 | 6.363E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.137E-05 | 0.01307505 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.451E-05 | 0.01112294 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.545E-05 | 8.886E-03 |
|---|
| somitogenesis | GO:0001756 |  | 1.851E-05 | 8.513E-03 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.11E-05 | 8.087E-03 |
|---|
| sex determination | GO:0007530 |  | 2.193E-05 | 7.206E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.575E-05 | 7.402E-03 |
|---|
| vasculogenesis | GO:0001570 |  | 3.464E-05 | 8.852E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 3.464E-05 | 7.967E-03 |
|---|
| epidermal growth factor receptor signaling pathway | GO:0007173 |  | 5.8E-05 | 0.01212754 |
|---|


