Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6918
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7488 | 3.846e-03 | 6.420e-04 | 7.830e-01 | 1.933e-06 |
|---|
| Loi | 0.2307 | 7.973e-02 | 1.122e-02 | 4.269e-01 | 3.818e-04 |
|---|
| Schmidt | 0.6763 | 0.000e+00 | 0.000e+00 | 4.190e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.423e-03 | 0.000e+00 |
|---|
| Wang | 0.2639 | 2.383e-03 | 4.106e-02 | 3.832e-01 | 3.749e-05 |
|---|
Expression data for subnetwork 6918 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
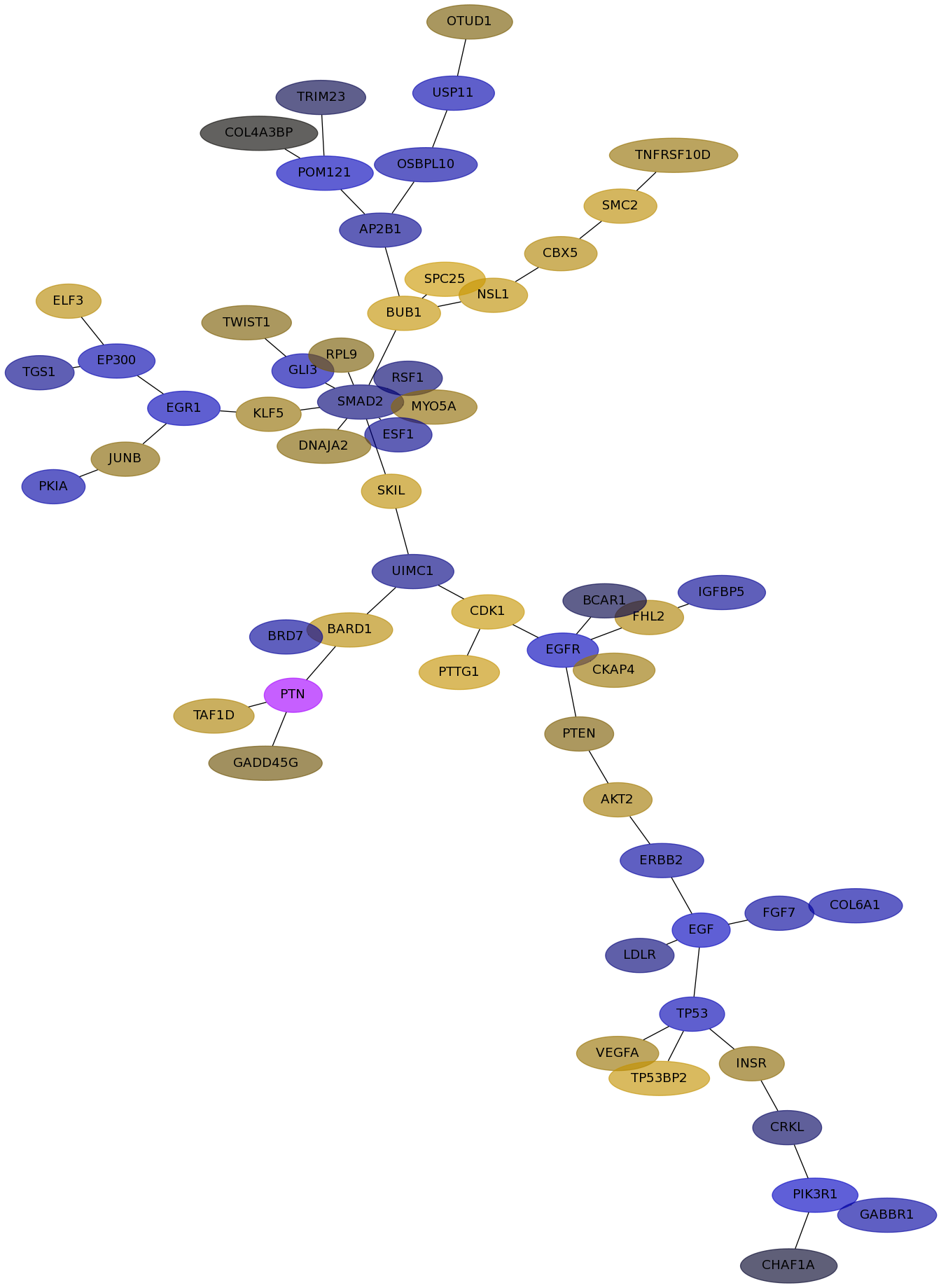
Score for each gene in subnetwork 6918 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| SPC25 |   | 1 | 1195 | 780 | 817 | 0.296 | -0.210 | 0.154 | 0.079 | -0.108 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| GABBR1 |   | 5 | 360 | 780 | 767 | -0.107 | 0.164 | 0.081 | 0.074 | 0.131 |
|---|
| EGF |   | 12 | 140 | 638 | 620 | -0.212 | 0.182 | -0.276 | -0.060 | 0.005 |
|---|
| LDLR |   | 5 | 360 | 780 | 767 | -0.041 | 0.218 | 0.039 | -0.028 | 0.064 |
|---|
| PTTG1 |   | 1 | 1195 | 780 | 817 | 0.251 | -0.139 | 0.036 | 0.001 | 0.128 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| RSF1 |   | 2 | 743 | 780 | 787 | -0.032 | 0.063 | 0.103 | 0.218 | 0.021 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| POM121 |   | 7 | 256 | 780 | 763 | -0.197 | 0.101 | 0.079 | undef | 0.017 |
|---|
| ELF3 |   | 1 | 1195 | 780 | 817 | 0.178 | -0.022 | -0.052 | 0.048 | -0.076 |
|---|
| CRKL |   | 6 | 301 | 780 | 764 | -0.023 | 0.056 | 0.271 | -0.025 | -0.008 |
|---|
| OTUD1 |   | 1 | 1195 | 780 | 817 | 0.041 | -0.105 | undef | undef | undef |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| COL6A1 |   | 5 | 360 | 780 | 767 | -0.112 | 0.193 | -0.013 | undef | 0.039 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| AP2B1 |   | 6 | 301 | 780 | 764 | -0.070 | -0.106 | 0.070 | 0.252 | 0.094 |
|---|
| BUB1 |   | 6 | 301 | 590 | 582 | 0.246 | -0.172 | 0.013 | -0.007 | 0.170 |
|---|
| ESF1 |   | 1 | 1195 | 780 | 817 | -0.064 | 0.070 | 0.098 | -0.004 | 0.200 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| MYO5A |   | 1 | 1195 | 780 | 817 | 0.071 | -0.021 | -0.066 | -0.082 | 0.135 |
|---|
| CKAP4 |   | 6 | 301 | 780 | 764 | 0.093 | 0.152 | -0.167 | 0.315 | -0.017 |
|---|
| TRIM23 |   | 1 | 1195 | 780 | 817 | -0.013 | -0.004 | 0.053 | -0.041 | 0.183 |
|---|
| RPL9 |   | 1 | 1195 | 780 | 817 | 0.041 | 0.112 | 0.158 | -0.284 | -0.041 |
|---|
| TAF1D |   | 10 | 167 | 590 | 566 | 0.141 | 0.113 | 0.041 | undef | 0.059 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| COL4A3BP |   | 1 | 1195 | 780 | 817 | 0.001 | -0.002 | 0.035 | 0.005 | -0.048 |
|---|
| OSBPL10 |   | 2 | 743 | 780 | 787 | -0.116 | 0.239 | 0.026 | 0.187 | 0.035 |
|---|
| SMC2 |   | 4 | 440 | 83 | 112 | 0.202 | -0.108 | 0.119 | 0.132 | 0.079 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| BCAR1 |   | 6 | 301 | 1 | 52 | -0.012 | 0.229 | undef | 0.223 | undef |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| CHAF1A |   | 1 | 1195 | 780 | 817 | -0.006 | -0.147 | 0.022 | -0.167 | -0.057 |
|---|
| SKIL |   | 3 | 557 | 780 | 781 | 0.209 | 0.108 | 0.073 | 0.094 | -0.020 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| SMAD2 |   | 4 | 440 | 590 | 585 | -0.039 | -0.199 | 0.076 | -0.003 | 0.247 |
|---|
| NSL1 |   | 2 | 743 | 780 | 787 | 0.221 | -0.079 | 0.288 | undef | 0.174 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| UIMC1 |   | 6 | 301 | 179 | 205 | -0.054 | 0.226 | 0.031 | -0.005 | 0.189 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| USP11 |   | 7 | 256 | 478 | 462 | -0.144 | 0.106 | 0.121 | 0.218 | -0.044 |
|---|
| TNFRSF10D |   | 1 | 1195 | 780 | 817 | 0.081 | 0.020 | 0.000 | -0.141 | undef |
|---|
| CBX5 |   | 5 | 360 | 1 | 56 | 0.155 | 0.016 | 0.148 | 0.030 | -0.009 |
|---|
| TGS1 |   | 5 | 360 | 780 | 767 | -0.060 | -0.044 | 0.232 | undef | 0.269 |
|---|
| DNAJA2 |   | 3 | 557 | 780 | 781 | 0.054 | 0.063 | -0.152 | -0.019 | 0.081 |
|---|
| PKIA |   | 11 | 148 | 780 | 759 | -0.120 | 0.085 | 0.249 | 0.164 | 0.044 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
GO Enrichment output for subnetwork 6918 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 6.083E-11 | 1.486E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.518E-09 | 1.855E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 4.54E-09 | 3.697E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.056E-08 | 6.449E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.105E-08 | 1.028E-05 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 6.273E-08 | 2.554E-05 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 9.826E-08 | 3.429E-05 |
|---|
| endothelial cell migration | GO:0043542 |  | 6.948E-07 | 2.122E-04 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 6.948E-07 | 1.886E-04 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.124E-06 | 2.746E-04 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 1.492E-06 | 3.315E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.122E-10 | 2.7E-07 |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 2.293E-09 | 2.759E-06 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 6.855E-09 | 5.498E-06 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.594E-08 | 9.586E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.176E-08 | 1.528E-05 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 9.458E-08 | 3.793E-05 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.481E-07 | 5.09E-05 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.045E-06 | 3.143E-04 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 1.045E-06 | 2.793E-04 |
|---|
| endothelial cell migration | GO:0043542 |  | 1.045E-06 | 2.514E-04 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 2.46E-06 | 5.381E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.233E-07 | 5.135E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.912E-06 | 2.199E-03 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 3.808E-06 | 2.92E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.636E-06 | 3.816E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.057E-05 | 4.863E-03 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.057E-05 | 4.053E-03 |
|---|
| angiogenesis | GO:0001525 |  | 1.943E-05 | 6.384E-03 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.246E-05 | 6.458E-03 |
|---|
| mammary gland development | GO:0030879 |  | 3.045E-05 | 7.781E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.977E-05 | 0.01144647 |
|---|
| inositol metabolic process | GO:0006020 |  | 6.699E-05 | 0.01400706 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.792E-07 | 3.302E-04 |
|---|
| regulation of cell migration | GO:0030334 |  | 2.609E-07 | 2.404E-04 |
|---|
| regulation of locomotion | GO:0040012 |  | 5.475E-07 | 3.363E-04 |
|---|
| regulation of cell motion | GO:0051270 |  | 5.475E-07 | 2.523E-04 |
|---|
| heart development | GO:0007507 |  | 1.063E-06 | 3.917E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.756E-06 | 8.465E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 4.149E-06 | 1.092E-03 |
|---|
| mammary gland development | GO:0030879 |  | 8.047E-06 | 1.854E-03 |
|---|
| gland development | GO:0048732 |  | 1.311E-05 | 2.684E-03 |
|---|
| anterior/posterior pattern formation | GO:0009952 |  | 1.449E-05 | 2.671E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.669E-05 | 2.797E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 2.233E-07 | 5.135E-04 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 1.912E-06 | 2.199E-03 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 3.808E-06 | 2.92E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 6.636E-06 | 3.816E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.057E-05 | 4.863E-03 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.057E-05 | 4.053E-03 |
|---|
| angiogenesis | GO:0001525 |  | 1.943E-05 | 6.384E-03 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 2.246E-05 | 6.458E-03 |
|---|
| mammary gland development | GO:0030879 |  | 3.045E-05 | 7.781E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 4.977E-05 | 0.01144647 |
|---|
| inositol metabolic process | GO:0006020 |  | 6.699E-05 | 0.01400706 |
|---|



