Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6903
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7490 | 3.834e-03 | 6.390e-04 | 7.826e-01 | 1.917e-06 |
|---|
| Loi | 0.2306 | 7.976e-02 | 1.123e-02 | 4.270e-01 | 3.823e-04 |
|---|
| Schmidt | 0.6763 | 0.000e+00 | 0.000e+00 | 4.186e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.426e-03 | 0.000e+00 |
|---|
| Wang | 0.2637 | 2.406e-03 | 4.128e-02 | 3.843e-01 | 3.816e-05 |
|---|
Expression data for subnetwork 6903 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
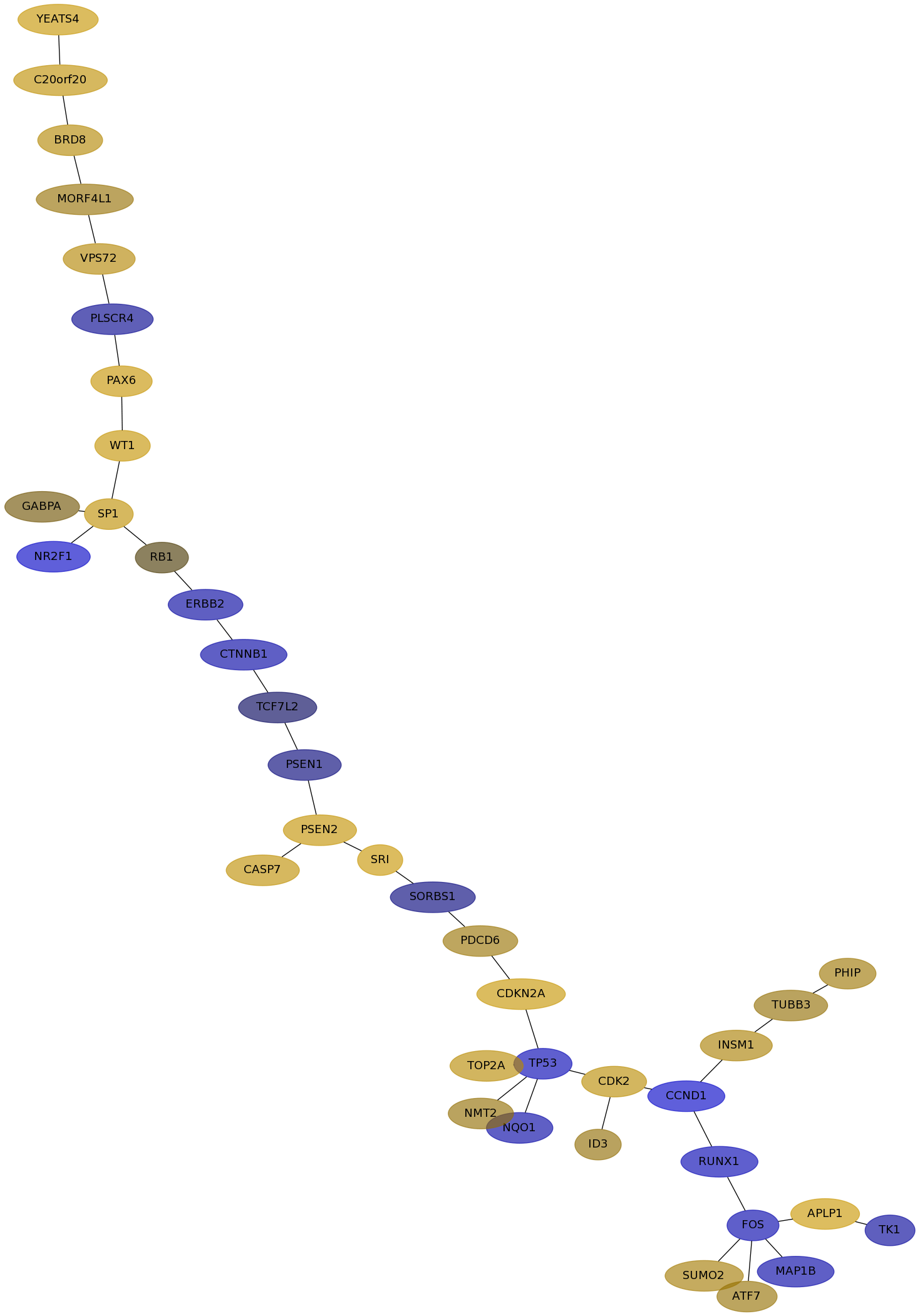
Score for each gene in subnetwork 6903 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| NQO1 |   | 21 | 73 | 366 | 354 | -0.115 | 0.069 | -0.189 | 0.321 | 0.118 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PSEN1 |   | 10 | 167 | 366 | 362 | -0.041 | 0.110 | undef | 0.083 | undef |
|---|
| TOP2A |   | 20 | 81 | 1 | 28 | 0.193 | -0.168 | 0.054 | -0.064 | 0.071 |
|---|
| MORF4L1 |   | 1 | 1195 | 906 | 946 | 0.081 | 0.104 | 0.315 | -0.013 | 0.061 |
|---|
| C20orf20 |   | 2 | 743 | 906 | 911 | 0.221 | -0.160 | 0.161 | 0.018 | 0.024 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| APLP1 |   | 8 | 222 | 727 | 703 | 0.284 | 0.043 | -0.105 | -0.121 | -0.190 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| BRD8 |   | 1 | 1195 | 906 | 946 | 0.172 | 0.151 | 0.134 | -0.122 | 0.007 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| VPS72 |   | 6 | 301 | 318 | 330 | 0.164 | -0.182 | 0.095 | 0.151 | 0.080 |
|---|
| SRI |   | 7 | 256 | 614 | 601 | 0.270 | -0.040 | 0.181 | 0.185 | 0.057 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| TUBB3 |   | 7 | 256 | 764 | 756 | 0.078 | 0.130 | 0.073 | 0.207 | -0.079 |
|---|
| PHIP |   | 4 | 440 | 906 | 902 | 0.099 | 0.100 | 0.156 | 0.136 | 0.042 |
|---|
| TK1 |   | 4 | 440 | 727 | 723 | -0.089 | -0.156 | 0.029 | 0.119 | -0.052 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| NMT2 |   | 7 | 256 | 906 | 876 | 0.077 | -0.296 | 0.134 | -0.124 | 0.217 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| ID3 |   | 9 | 196 | 658 | 637 | 0.072 | -0.006 | 0.192 | -0.023 | 0.057 |
|---|
| YEATS4 |   | 1 | 1195 | 906 | 946 | 0.268 | -0.032 | 0.120 | -0.062 | 0.050 |
|---|
| PDCD6 |   | 10 | 167 | 1 | 40 | 0.091 | -0.060 | undef | undef | undef |
|---|
| CASP7 |   | 6 | 301 | 614 | 602 | 0.216 | 0.095 | 0.255 | undef | 0.035 |
|---|
| RUNX1 |   | 5 | 360 | 906 | 890 | -0.161 | 0.140 | 0.016 | 0.102 | 0.025 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
GO Enrichment output for subnetwork 6903 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 8.865E-11 | 2.166E-07 |
|---|
| positive regulation of myeloid leukocyte differentiation | GO:0002763 |  | 8.615E-09 | 1.052E-05 |
|---|
| Notch receptor processing | GO:0007220 |  | 2.423E-07 | 1.973E-04 |
|---|
| pancreas development | GO:0031016 |  | 3.232E-07 | 1.974E-04 |
|---|
| positive regulation of myeloid cell differentiation | GO:0045639 |  | 3.232E-07 | 1.579E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 4.836E-07 | 1.969E-04 |
|---|
| regulation of myeloid leukocyte differentiation | GO:0002761 |  | 4.857E-07 | 1.695E-04 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 8.444E-07 | 2.579E-04 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.341E-06 | 3.641E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 1.348E-06 | 3.294E-04 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.348E-06 | 2.994E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.268E-11 | 1.508E-07 |
|---|
| Notch receptor processing | GO:0007220 |  | 2.838E-07 | 3.414E-04 |
|---|
| positive regulation of myeloid cell differentiation | GO:0045639 |  | 3.187E-07 | 2.556E-04 |
|---|
| pancreas development | GO:0031016 |  | 3.974E-07 | 2.39E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 5.663E-07 | 2.725E-04 |
|---|
| cell cycle arrest | GO:0007050 |  | 6.949E-07 | 2.787E-04 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 9.888E-07 | 3.399E-04 |
|---|
| fat cell differentiation | GO:0045444 |  | 1.416E-06 | 4.258E-04 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 1.578E-06 | 4.22E-04 |
|---|
| positive regulation of myeloid leukocyte differentiation | GO:0002763 |  | 1.578E-06 | 3.798E-04 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.578E-06 | 3.453E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of myeloid cell differentiation | GO:0045639 |  | 7.313E-07 | 1.682E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.972E-06 | 3.418E-03 |
|---|
| positive regulation of myeloid leukocyte differentiation | GO:0002763 |  | 2.972E-06 | 2.279E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 2.972E-06 | 1.709E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 6.334E-06 | 2.914E-03 |
|---|
| negative regulation of mast cell proliferation | GO:0070667 |  | 8.685E-06 | 3.329E-03 |
|---|
| regulation of myeloid cell differentiation | GO:0045637 |  | 1.623E-05 | 5.333E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.947E-05 | 5.596E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.947E-05 | 4.974E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.173E-05 | 4.997E-03 |
|---|
| interphase | GO:0051325 |  | 2.177E-05 | 4.551E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell growth | GO:0001558 |  | 5.84E-08 | 1.076E-04 |
|---|
| chromosome segregation | GO:0007059 |  | 2.892E-07 | 2.665E-04 |
|---|
| amyloid precursor protein catabolic process | GO:0042987 |  | 5.363E-07 | 3.295E-04 |
|---|
| amyloid precursor protein metabolic process | GO:0042982 |  | 1.867E-06 | 8.604E-04 |
|---|
| membrane protein ectodomain proteolysis | GO:0006509 |  | 1.867E-06 | 6.883E-04 |
|---|
| positive regulation of myeloid leukocyte differentiation | GO:0002763 |  | 2.98E-06 | 9.153E-04 |
|---|
| glycoprotein catabolic process | GO:0006516 |  | 1.158E-05 | 3.05E-03 |
|---|
| G1 phase | GO:0051318 |  | 1.158E-05 | 2.669E-03 |
|---|
| forebrain development | GO:0030900 |  | 1.902E-05 | 3.895E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.377E-05 | 4.381E-03 |
|---|
| positive regulation of myeloid cell differentiation | GO:0045639 |  | 2.377E-05 | 3.983E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of myeloid cell differentiation | GO:0045639 |  | 7.313E-07 | 1.682E-03 |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.972E-06 | 3.418E-03 |
|---|
| positive regulation of myeloid leukocyte differentiation | GO:0002763 |  | 2.972E-06 | 2.279E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 2.972E-06 | 1.709E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 6.334E-06 | 2.914E-03 |
|---|
| negative regulation of mast cell proliferation | GO:0070667 |  | 8.685E-06 | 3.329E-03 |
|---|
| regulation of myeloid cell differentiation | GO:0045637 |  | 1.623E-05 | 5.333E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.947E-05 | 5.596E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.947E-05 | 4.974E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.173E-05 | 4.997E-03 |
|---|
| interphase | GO:0051325 |  | 2.177E-05 | 4.551E-03 |
|---|



