Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6864
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7490 | 3.836e-03 | 6.400e-04 | 7.827e-01 | 1.922e-06 |
|---|
| Loi | 0.2310 | 7.923e-02 | 1.109e-02 | 4.257e-01 | 3.740e-04 |
|---|
| Schmidt | 0.6759 | 0.000e+00 | 0.000e+00 | 4.226e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.429e-03 | 0.000e+00 |
|---|
| Wang | 0.2635 | 2.429e-03 | 4.150e-02 | 3.853e-01 | 3.884e-05 |
|---|
Expression data for subnetwork 6864 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
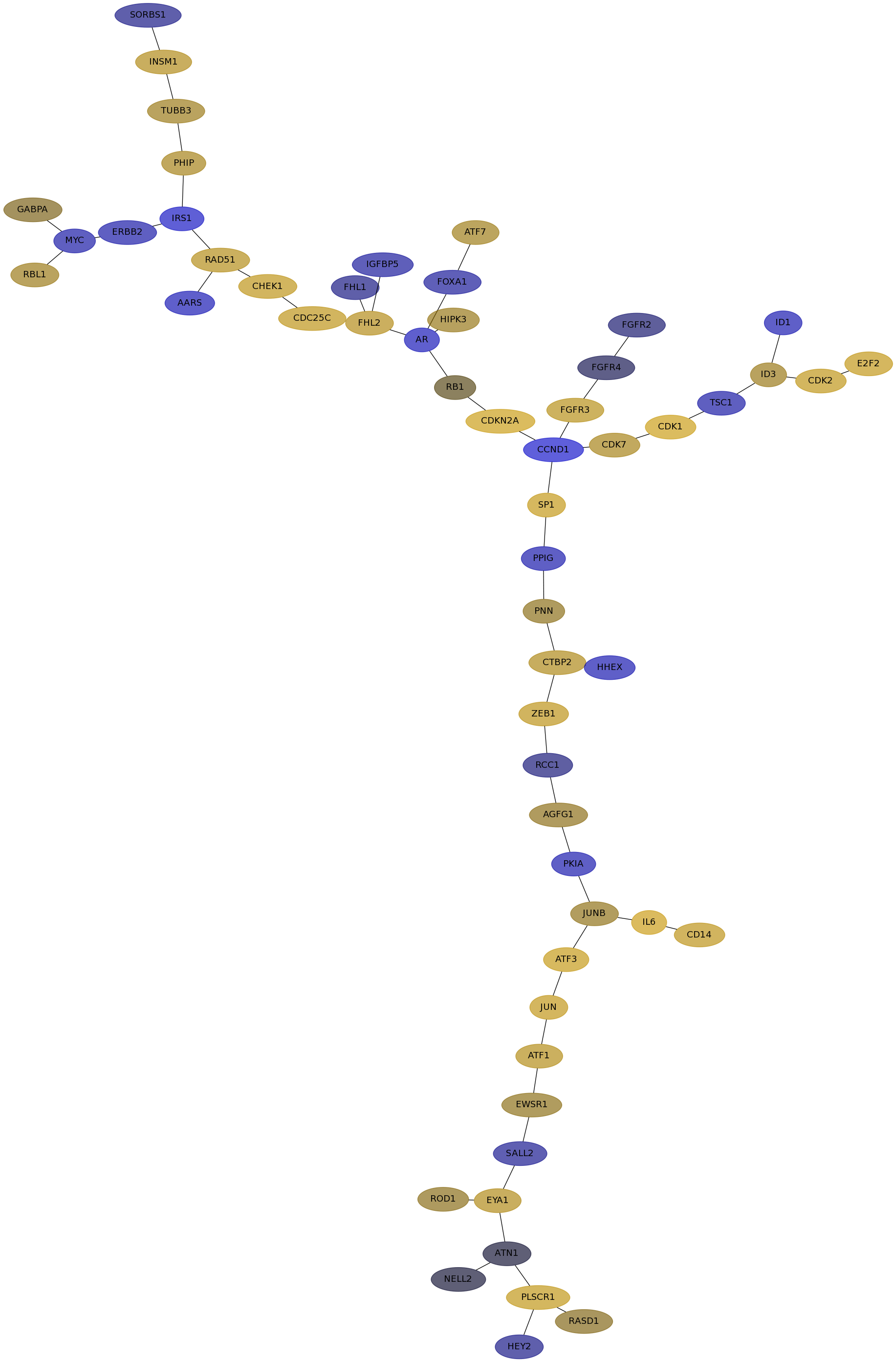
Score for each gene in subnetwork 6864 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EWSR1 |   | 1 | 1195 | 1564 | 1578 | 0.054 | 0.150 | 0.161 | 0.134 | -0.010 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| ID1 |   | 3 | 557 | 806 | 792 | -0.128 | -0.030 | 0.124 | 0.069 | -0.142 |
|---|
| EYA1 |   | 8 | 222 | 1010 | 981 | 0.135 | -0.065 | 0.162 | 0.068 | -0.088 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| RASD1 |   | 1 | 1195 | 1564 | 1578 | 0.040 | -0.001 | undef | -0.015 | undef |
|---|
| CTBP2 |   | 1 | 1195 | 1564 | 1578 | 0.125 | 0.141 | 0.183 | undef | -0.183 |
|---|
| AGFG1 |   | 1 | 1195 | 1564 | 1578 | 0.052 | -0.069 | -0.184 | 0.094 | 0.057 |
|---|
| AARS |   | 2 | 743 | 1564 | 1549 | -0.148 | 0.030 | -0.164 | 0.177 | 0.035 |
|---|
| SALL2 |   | 1 | 1195 | 1564 | 1578 | -0.058 | 0.015 | 0.209 | undef | -0.058 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| HEY2 |   | 8 | 222 | 570 | 555 | -0.045 | -0.021 | 0.398 | -0.024 | -0.058 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| FGFR3 |   | 9 | 196 | 179 | 195 | 0.160 | 0.071 | 0.018 | -0.050 | -0.111 |
|---|
| ATF1 |   | 1 | 1195 | 1564 | 1578 | 0.146 | 0.007 | 0.098 | -0.075 | 0.221 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| NELL2 |   | 7 | 256 | 727 | 714 | -0.005 | 0.138 | 0.421 | -0.027 | 0.074 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| PPIG |   | 8 | 222 | 648 | 631 | -0.116 | 0.024 | 0.221 | -0.065 | 0.101 |
|---|
| JUNB |   | 20 | 81 | 179 | 185 | 0.057 | 0.086 | 0.065 | 0.001 | -0.015 |
|---|
| ZEB1 |   | 2 | 743 | 1564 | 1549 | 0.178 | -0.158 | -0.094 | undef | -0.158 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| TUBB3 |   | 7 | 256 | 764 | 756 | 0.078 | 0.130 | 0.073 | 0.207 | -0.079 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| PHIP |   | 4 | 440 | 906 | 902 | 0.099 | 0.100 | 0.156 | 0.136 | 0.042 |
|---|
| PLSCR1 |   | 10 | 167 | 412 | 408 | 0.202 | -0.159 | 0.091 | undef | -0.105 |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| CD14 |   | 2 | 743 | 366 | 395 | 0.183 | -0.144 | 0.050 | -0.207 | 0.013 |
|---|
| CDK7 |   | 7 | 256 | 318 | 328 | 0.105 | 0.184 | 0.017 | 0.078 | -0.097 |
|---|
| PNN |   | 1 | 1195 | 1564 | 1578 | 0.051 | 0.175 | 0.218 | -0.037 | -0.186 |
|---|
| CDC25C |   | 3 | 557 | 658 | 654 | 0.189 | 0.029 | 0.201 | -0.055 | -0.062 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| IRS1 |   | 9 | 196 | 842 | 805 | -0.219 | 0.065 | 0.165 | 0.102 | 0.064 |
|---|
| TSC1 |   | 2 | 743 | 1564 | 1549 | -0.094 | 0.257 | 0.103 | -0.042 | 0.077 |
|---|
| ID3 |   | 9 | 196 | 658 | 637 | 0.072 | -0.006 | 0.192 | -0.023 | 0.057 |
|---|
| HHEX |   | 4 | 440 | 727 | 723 | -0.130 | -0.178 | 0.177 | -0.180 | 0.005 |
|---|
| RCC1 |   | 1 | 1195 | 1564 | 1578 | -0.031 | -0.030 | -0.057 | 0.010 | -0.151 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| FGFR2 |   | 7 | 256 | 179 | 201 | -0.024 | -0.001 | 0.129 | undef | 0.009 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| IL6 |   | 9 | 196 | 366 | 365 | 0.257 | -0.106 | 0.226 | 0.024 | 0.035 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| ATN1 |   | 7 | 256 | 412 | 414 | -0.005 | -0.093 | 0.005 | -0.185 | -0.136 |
|---|
| ROD1 |   | 4 | 440 | 1564 | 1542 | 0.049 | 0.088 | 0.052 | 0.011 | -0.223 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| HIPK3 |   | 9 | 196 | 1461 | 1417 | 0.069 | 0.005 | -0.027 | undef | 0.175 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| PKIA |   | 11 | 148 | 780 | 759 | -0.120 | 0.085 | 0.249 | 0.164 | 0.044 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6864 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.041E-10 | 9.873E-07 |
|---|
| viral genome replication | GO:0019079 |  | 5.058E-10 | 6.179E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.804E-09 | 3.912E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.162E-08 | 1.32E-05 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 2.162E-08 | 1.056E-05 |
|---|
| cell growth | GO:0016049 |  | 4.331E-08 | 1.763E-05 |
|---|
| viral infectious cycle | GO:0019058 |  | 8.182E-08 | 2.856E-05 |
|---|
| gland development | GO:0048732 |  | 1.304E-07 | 3.981E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 1.416E-07 | 3.844E-05 |
|---|
| viral reproductive process | GO:0022415 |  | 2.688E-07 | 6.566E-05 |
|---|
| viral reproduction | GO:0016032 |  | 4.686E-07 | 1.041E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.141E-10 | 9.964E-07 |
|---|
| interphase | GO:0051325 |  | 5.485E-10 | 6.598E-07 |
|---|
| viral genome replication | GO:0019079 |  | 6.283E-10 | 5.039E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.145E-09 | 3.696E-06 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 2.51E-08 | 1.208E-05 |
|---|
| cell growth | GO:0016049 |  | 4.682E-08 | 1.877E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 7.706E-08 | 2.649E-05 |
|---|
| viral infectious cycle | GO:0019058 |  | 1.013E-07 | 3.046E-05 |
|---|
| gland development | GO:0048732 |  | 1.317E-07 | 3.521E-05 |
|---|
| viral reproductive process | GO:0022415 |  | 3.323E-07 | 7.995E-05 |
|---|
| viral reproduction | GO:0016032 |  | 5.789E-07 | 1.266E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| viral genome replication | GO:0019079 |  | 2.97E-09 | 6.831E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.017E-08 | 5.77E-05 |
|---|
| interphase | GO:0051325 |  | 6.017E-08 | 4.613E-05 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 8.772E-08 | 5.044E-05 |
|---|
| cell growth | GO:0016049 |  | 1.195E-07 | 5.496E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.89E-07 | 1.108E-04 |
|---|
| viral infectious cycle | GO:0019058 |  | 3.781E-07 | 1.242E-04 |
|---|
| gland development | GO:0048732 |  | 4.186E-07 | 1.204E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.218E-07 | 1.589E-04 |
|---|
| viral reproductive process | GO:0022415 |  | 1.35E-06 | 3.106E-04 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 1.656E-06 | 3.462E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| gland development | GO:0048732 |  | 6.36E-09 | 1.172E-05 |
|---|
| response to insulin stimulus | GO:0032868 |  | 2.366E-07 | 2.18E-04 |
|---|
| response to hormone stimulus | GO:0009725 |  | 9.679E-07 | 5.946E-04 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 9.694E-07 | 4.466E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.034E-06 | 3.812E-04 |
|---|
| prostate gland development | GO:0030850 |  | 1.543E-06 | 4.74E-04 |
|---|
| endocrine pancreas development | GO:0031018 |  | 1.543E-06 | 4.063E-04 |
|---|
| meiosis I | GO:0007127 |  | 2.291E-06 | 5.278E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.074E-06 | 6.296E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.074E-06 | 5.666E-04 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 4.428E-06 | 7.419E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| viral genome replication | GO:0019079 |  | 2.97E-09 | 6.831E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 5.017E-08 | 5.77E-05 |
|---|
| interphase | GO:0051325 |  | 6.017E-08 | 4.613E-05 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 8.772E-08 | 5.044E-05 |
|---|
| cell growth | GO:0016049 |  | 1.195E-07 | 5.496E-05 |
|---|
| fibroblast growth factor receptor signaling pathway | GO:0008543 |  | 2.89E-07 | 1.108E-04 |
|---|
| viral infectious cycle | GO:0019058 |  | 3.781E-07 | 1.242E-04 |
|---|
| gland development | GO:0048732 |  | 4.186E-07 | 1.204E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.218E-07 | 1.589E-04 |
|---|
| viral reproductive process | GO:0022415 |  | 1.35E-06 | 3.106E-04 |
|---|
| regulation of protein ubiquitination | GO:0031396 |  | 1.656E-06 | 3.462E-04 |
|---|



