Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6852
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7491 | 3.829e-03 | 6.390e-04 | 7.825e-01 | 1.914e-06 |
|---|
| Loi | 0.2308 | 7.949e-02 | 1.116e-02 | 4.263e-01 | 3.781e-04 |
|---|
| Schmidt | 0.6758 | 0.000e+00 | 0.000e+00 | 4.238e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.424e-03 | 0.000e+00 |
|---|
| Wang | 0.2635 | 2.425e-03 | 4.146e-02 | 3.851e-01 | 3.872e-05 |
|---|
Expression data for subnetwork 6852 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
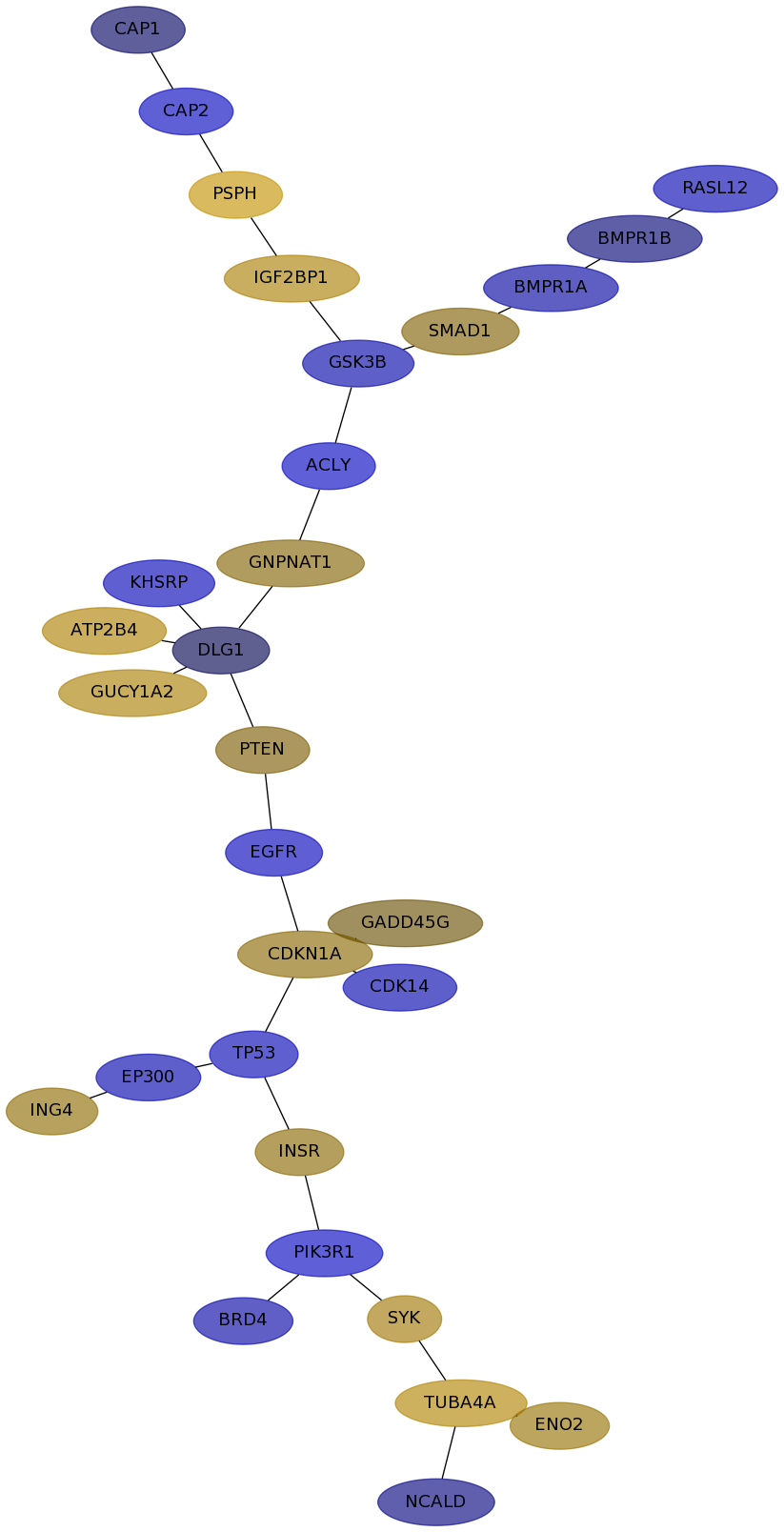
Score for each gene in subnetwork 6852 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| BRD4 |   | 11 | 148 | 1047 | 1003 | -0.119 | 0.095 | -0.075 | 0.081 | -0.044 |
|---|
| ENO2 |   | 3 | 557 | 727 | 728 | 0.085 | 0.114 | 0.148 | -0.071 | 0.208 |
|---|
| GNPNAT1 |   | 2 | 743 | 1260 | 1256 | 0.054 | -0.034 | undef | 0.150 | undef |
|---|
| ING4 |   | 1 | 1195 | 1709 | 1725 | 0.070 | 0.012 | -0.108 | -0.001 | 0.050 |
|---|
| CAP2 |   | 2 | 743 | 570 | 593 | -0.214 | -0.017 | 0.206 | 0.196 | 0.169 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| GUCY1A2 |   | 2 | 743 | 457 | 481 | 0.135 | -0.058 | -0.088 | 0.307 | 0.131 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| CDK14 |   | 7 | 256 | 179 | 201 | -0.151 | 0.011 | -0.071 | 0.212 | 0.107 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CAP1 |   | 1 | 1195 | 1709 | 1725 | -0.025 | 0.189 | -0.047 | undef | 0.098 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| NCALD |   | 4 | 440 | 1709 | 1672 | -0.046 | 0.078 | 0.090 | undef | 0.004 |
|---|
| ACLY |   | 6 | 301 | 1260 | 1224 | -0.236 | 0.071 | 0.009 | 0.158 | 0.115 |
|---|
| DLG1 |   | 6 | 301 | 457 | 452 | -0.016 | 0.173 | 0.244 | 0.017 | 0.123 |
|---|
| KHSRP |   | 2 | 743 | 1709 | 1695 | -0.192 | 0.131 | -0.114 | -0.045 | 0.144 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| BMPR1A |   | 1 | 1195 | 1709 | 1725 | -0.108 | 0.122 | 0.140 | 0.038 | -0.064 |
|---|
| SMAD1 |   | 5 | 360 | 682 | 671 | 0.049 | 0.159 | -0.062 | 0.140 | 0.048 |
|---|
| SYK |   | 2 | 743 | 1510 | 1502 | 0.106 | -0.251 | -0.134 | 0.017 | 0.163 |
|---|
| ATP2B4 |   | 1 | 1195 | 1709 | 1725 | 0.142 | 0.177 | -0.004 | undef | 0.070 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| RASL12 |   | 1 | 1195 | 1709 | 1725 | -0.154 | 0.210 | 0.152 | -0.042 | 0.004 |
|---|
| TUBA4A |   | 7 | 256 | 1 | 47 | 0.157 | 0.136 | -0.010 | undef | 0.156 |
|---|
| BMPR1B |   | 2 | 743 | 1709 | 1695 | -0.037 | 0.078 | 0.163 | -0.003 | 0.146 |
|---|
GO Enrichment output for subnetwork 6852 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 1.379E-10 | 3.37E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 4.131E-10 | 5.046E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 9.621E-10 | 7.835E-07 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.921E-09 | 1.173E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.45E-09 | 2.174E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 5.742E-09 | 2.338E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 9.007E-09 | 3.143E-06 |
|---|
| endothelial cell migration | GO:0043542 |  | 6.427E-08 | 1.963E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 6.427E-08 | 1.745E-05 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 6.427E-08 | 1.57E-05 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.043E-07 | 2.316E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 2.188E-10 | 5.263E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 6.55E-10 | 7.879E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.525E-09 | 1.223E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.044E-09 | 1.831E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.757E-09 | 4.214E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 9.097E-09 | 3.648E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.427E-08 | 4.904E-06 |
|---|
| endothelial cell migration | GO:0043542 |  | 1.017E-07 | 3.058E-05 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.017E-07 | 2.718E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.017E-07 | 2.446E-05 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 1.017E-07 | 2.224E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.62E-07 | 6.026E-04 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 3.518E-07 | 4.046E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.461E-07 | 3.42E-04 |
|---|
| ER overload response | GO:0006983 |  | 5.229E-07 | 3.006E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 5.229E-07 | 2.405E-04 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 7.647E-07 | 2.931E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 9.13E-07 | 3E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.458E-06 | 4.191E-04 |
|---|
| BMP signaling pathway | GO:0030509 |  | 1.46E-06 | 3.732E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 3.11E-06 | 7.154E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.11E-06 | 6.503E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 9.295E-08 | 1.713E-04 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 2.195E-07 | 2.023E-04 |
|---|
| BMP signaling pathway | GO:0030509 |  | 2.816E-07 | 1.73E-04 |
|---|
| negative regulation of cell cycle | GO:0045786 |  | 3.82E-07 | 1.76E-04 |
|---|
| ER overload response | GO:0006983 |  | 6.486E-07 | 2.391E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.843E-06 | 5.662E-04 |
|---|
| regulation of ossification | GO:0030278 |  | 2.456E-06 | 6.467E-04 |
|---|
| positive regulation of biomineral formation | GO:0070169 |  | 3.858E-06 | 8.887E-04 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 4.12E-06 | 8.436E-04 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 4.641E-06 | 8.553E-04 |
|---|
| protein amino acid acetylation | GO:0006473 |  | 5.293E-06 | 8.868E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 2.62E-07 | 6.026E-04 |
|---|
| positive regulation of osteoblast differentiation | GO:0045669 |  | 3.518E-07 | 4.046E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.461E-07 | 3.42E-04 |
|---|
| ER overload response | GO:0006983 |  | 5.229E-07 | 3.006E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 5.229E-07 | 2.405E-04 |
|---|
| regulation of osteoblast differentiation | GO:0045667 |  | 7.647E-07 | 2.931E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 9.13E-07 | 3E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 1.458E-06 | 4.191E-04 |
|---|
| BMP signaling pathway | GO:0030509 |  | 1.46E-06 | 3.732E-04 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 3.11E-06 | 7.154E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 3.11E-06 | 6.503E-04 |
|---|



