Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6836
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7495 | 3.799e-03 | 6.320e-04 | 7.814e-01 | 1.876e-06 |
|---|
| Loi | 0.2308 | 7.946e-02 | 1.115e-02 | 4.262e-01 | 3.775e-04 |
|---|
| Schmidt | 0.6756 | 0.000e+00 | 0.000e+00 | 4.258e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.424e-03 | 0.000e+00 |
|---|
| Wang | 0.2636 | 2.421e-03 | 4.142e-02 | 3.850e-01 | 3.860e-05 |
|---|
Expression data for subnetwork 6836 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
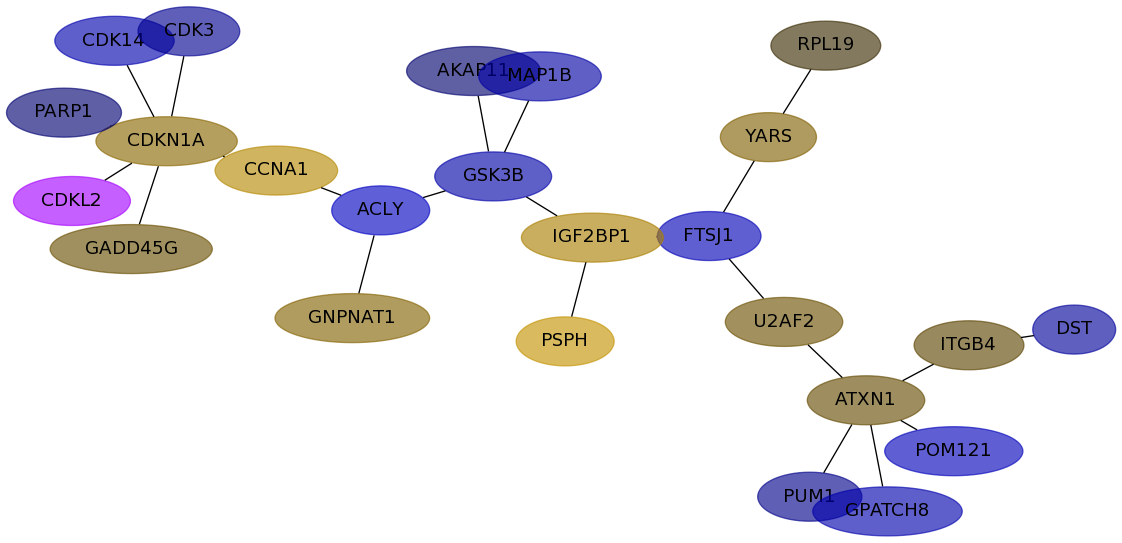
Score for each gene in subnetwork 6836 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| U2AF2 |   | 2 | 743 | 1260 | 1256 | 0.031 | 0.113 | 0.171 | -0.075 | 0.082 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| GNPNAT1 |   | 2 | 743 | 1260 | 1256 | 0.054 | -0.034 | undef | 0.150 | undef |
|---|
| PUM1 |   | 1 | 1195 | 1260 | 1282 | -0.065 | 0.205 | 0.172 | undef | -0.009 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| CDKL2 |   | 3 | 557 | 1260 | 1237 | undef | -0.129 | 0.000 | 0.109 | undef |
|---|
| POM121 |   | 7 | 256 | 780 | 763 | -0.197 | 0.101 | 0.079 | undef | 0.017 |
|---|
| CDK3 |   | 7 | 256 | 590 | 570 | -0.076 | 0.189 | -0.041 | -0.089 | 0.082 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| CDK14 |   | 7 | 256 | 179 | 201 | -0.151 | 0.011 | -0.071 | 0.212 | 0.107 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| GPATCH8 |   | 8 | 222 | 614 | 599 | -0.143 | 0.014 | 0.019 | -0.070 | 0.061 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| ACLY |   | 6 | 301 | 1260 | 1224 | -0.236 | 0.071 | 0.009 | 0.158 | 0.115 |
|---|
| FTSJ1 |   | 1 | 1195 | 1260 | 1282 | -0.178 | -0.023 | -0.082 | 0.177 | 0.212 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| RPL19 |   | 1 | 1195 | 1260 | 1282 | 0.009 | 0.043 | -0.022 | 0.050 | 0.063 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| YARS |   | 1 | 1195 | 1260 | 1282 | 0.051 | 0.100 | 0.033 | 0.097 | 0.057 |
|---|
| AKAP11 |   | 4 | 440 | 457 | 458 | -0.034 | -0.099 | -0.038 | 0.251 | 0.017 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
GO Enrichment output for subnetwork 6836 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 8.657E-08 | 2.115E-04 |
|---|
| L-serine metabolic process | GO:0006563 |  | 7.1E-07 | 8.673E-04 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 1.561E-06 | 1.271E-03 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 8.577E-06 | 5.238E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.889E-05 | 9.23E-03 |
|---|
| serine family amino acid metabolic process | GO:0009069 |  | 2.506E-05 | 0.01020354 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 3.242E-05 | 0.01131607 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 4.046E-05 | 0.01235453 |
|---|
| cytoplasmic microtubule organization | GO:0031122 |  | 4.046E-05 | 0.01098181 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.658E-05 | 0.01382229 |
|---|
| rRNA modification | GO:0000154 |  | 5.658E-05 | 0.01256572 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 5.133E-08 | 1.235E-04 |
|---|
| L-serine metabolic process | GO:0006563 |  | 6.121E-07 | 7.364E-04 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 1.453E-06 | 1.166E-03 |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 7.739E-06 | 4.655E-03 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 1.82E-05 | 8.76E-03 |
|---|
| serine family amino acid metabolic process | GO:0009069 |  | 2.234E-05 | 8.958E-03 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 3.236E-05 | 0.01112406 |
|---|
| male meiosis | GO:0007140 |  | 4.537E-05 | 0.01364599 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 4.537E-05 | 0.01212977 |
|---|
| cytoplasmic microtubule organization | GO:0031122 |  | 4.537E-05 | 0.01091679 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 6.345E-05 | 0.01387842 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 1.048E-05 | 0.02411156 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 2.782E-05 | 0.03199044 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 4.316E-05 | 0.0330922 |
|---|
| male meiosis | GO:0007140 |  | 7.613E-05 | 0.04377311 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.613E-05 | 0.03501848 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 7.613E-05 | 0.02918207 |
|---|
| ER overload response | GO:0006983 |  | 7.613E-05 | 0.0250132 |
|---|
| coenzyme A metabolic process | GO:0015936 |  | 7.613E-05 | 0.02188655 |
|---|
| RNA methylation | GO:0001510 |  | 1.064E-04 | 0.02719715 |
|---|
| microtubule bundle formation | GO:0001578 |  | 1.064E-04 | 0.02447743 |
|---|
| rRNA modification | GO:0000154 |  | 1.064E-04 | 0.02225221 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| ATP catabolic process | GO:0006200 |  | 6.436E-05 | 0.1186072 |
|---|
| ER overload response | GO:0006983 |  | 9.638E-05 | 0.08881748 |
|---|
| rRNA modification | GO:0000154 |  | 9.638E-05 | 0.05921165 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 9.638E-05 | 0.04440874 |
|---|
| RNA methylation | GO:0001510 |  | 9.638E-05 | 0.03552699 |
|---|
| ribonucleoside triphosphate catabolic process | GO:0009203 |  | 9.638E-05 | 0.02960583 |
|---|
| L-serine metabolic process | GO:0006563 |  | 9.638E-05 | 0.02537642 |
|---|
| peptidyl-serine phosphorylation | GO:0018105 |  | 9.638E-05 | 0.02220437 |
|---|
| integrin-mediated signaling pathway | GO:0007229 |  | 1.017E-04 | 0.02083435 |
|---|
| axon cargo transport | GO:0008088 |  | 1.347E-04 | 0.02483034 |
|---|
| purine nucleoside triphosphate catabolic process | GO:0009146 |  | 1.347E-04 | 0.02257304 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of microtubule polymerization or depolymerization | GO:0031110 |  | 1.048E-05 | 0.02411156 |
|---|
| regulation of microtubule cytoskeleton organization | GO:0070507 |  | 2.782E-05 | 0.03199044 |
|---|
| regulation of microtubule-based process | GO:0032886 |  | 4.316E-05 | 0.0330922 |
|---|
| male meiosis | GO:0007140 |  | 7.613E-05 | 0.04377311 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.613E-05 | 0.03501848 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 7.613E-05 | 0.02918207 |
|---|
| ER overload response | GO:0006983 |  | 7.613E-05 | 0.0250132 |
|---|
| coenzyme A metabolic process | GO:0015936 |  | 7.613E-05 | 0.02188655 |
|---|
| RNA methylation | GO:0001510 |  | 1.064E-04 | 0.02719715 |
|---|
| microtubule bundle formation | GO:0001578 |  | 1.064E-04 | 0.02447743 |
|---|
| rRNA modification | GO:0000154 |  | 1.064E-04 | 0.02225221 |
|---|



