Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6821
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7496 | 3.794e-03 | 6.310e-04 | 7.812e-01 | 1.870e-06 |
|---|
| Loi | 0.2307 | 7.962e-02 | 1.119e-02 | 4.266e-01 | 3.801e-04 |
|---|
| Schmidt | 0.6757 | 0.000e+00 | 0.000e+00 | 4.250e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7911 | 0.000e+00 | 0.000e+00 | 3.426e-03 | 0.000e+00 |
|---|
| Wang | 0.2635 | 2.427e-03 | 4.147e-02 | 3.852e-01 | 3.877e-05 |
|---|
Expression data for subnetwork 6821 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
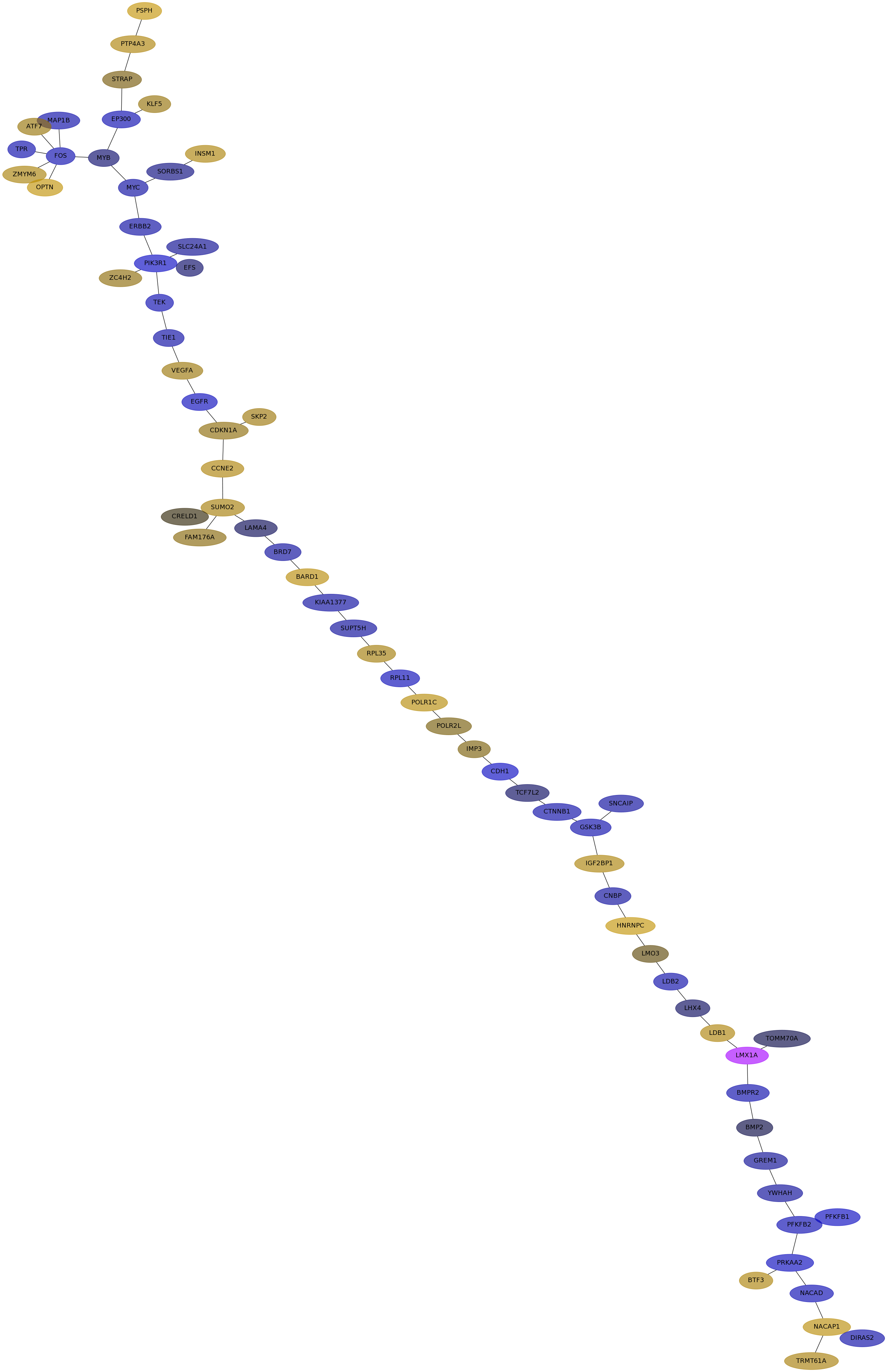
Score for each gene in subnetwork 6821 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| TEK |   | 18 | 86 | 83 | 95 | -0.149 | -0.049 | 0.174 | 0.015 | 0.113 |
|---|
| CCNE2 |   | 11 | 148 | 412 | 407 | 0.136 | -0.178 | 0.241 | undef | -0.055 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| SUMO2 |   | 22 | 69 | 366 | 345 | 0.117 | 0.062 | 0.294 | 0.141 | -0.096 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PTP4A3 |   | 9 | 196 | 412 | 411 | 0.137 | -0.196 | 0.085 | undef | -0.004 |
|---|
| EFS |   | 3 | 557 | 1323 | 1309 | -0.024 | 0.300 | -0.003 | 0.208 | -0.034 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| CRELD1 |   | 2 | 743 | 236 | 272 | 0.006 | 0.130 | 0.075 | 0.111 | 0.095 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| RPL11 |   | 1 | 1195 | 1323 | 1347 | -0.176 | 0.076 | 0.295 | -0.325 | -0.047 |
|---|
| ZMYM6 |   | 1 | 1195 | 1323 | 1347 | 0.126 | -0.218 | 0.092 | 0.240 | 0.055 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| DIRAS2 |   | 1 | 1195 | 1323 | 1347 | -0.113 | 0.072 | 0.164 | 0.045 | -0.005 |
|---|
| STRAP |   | 7 | 256 | 412 | 414 | 0.035 | -0.128 | 0.074 | 0.154 | -0.049 |
|---|
| LAMA4 |   | 13 | 124 | 696 | 670 | -0.016 | 0.138 | 0.209 | 0.172 | 0.343 |
|---|
| LMO3 |   | 1 | 1195 | 1323 | 1347 | 0.020 | -0.068 | -0.050 | 0.138 | -0.092 |
|---|
| RPL35 |   | 8 | 222 | 1323 | 1280 | 0.108 | -0.012 | 0.370 | -0.145 | -0.220 |
|---|
| FAM176A |   | 2 | 743 | 1116 | 1121 | 0.054 | 0.217 | undef | undef | undef |
|---|
| POLR2L |   | 5 | 360 | 727 | 722 | 0.037 | -0.012 | 0.224 | 0.054 | 0.015 |
|---|
| PFKFB2 |   | 3 | 557 | 692 | 691 | -0.149 | -0.081 | -0.158 | 0.121 | -0.202 |
|---|
| HNRNPC |   | 1 | 1195 | 1323 | 1347 | 0.237 | 0.062 | 0.295 | undef | 0.037 |
|---|
| LDB2 |   | 1 | 1195 | 1323 | 1347 | -0.118 | 0.055 | 0.122 | undef | 0.078 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| OPTN |   | 7 | 256 | 1 | 47 | 0.226 | -0.099 | 0.015 | undef | 0.035 |
|---|
| SLC24A1 |   | 4 | 440 | 1323 | 1295 | -0.068 | 0.109 | 0.178 | -0.020 | 0.056 |
|---|
| LHX4 |   | 1 | 1195 | 1323 | 1347 | -0.020 | 0.112 | undef | undef | undef |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| BTF3 |   | 1 | 1195 | 1323 | 1347 | 0.135 | 0.057 | 0.100 | 0.054 | -0.032 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| PRKAA2 |   | 6 | 301 | 1165 | 1138 | -0.204 | 0.148 | 0.086 | 0.250 | -0.078 |
|---|
| NACAP1 |   | 2 | 743 | 1323 | 1313 | 0.186 | 0.227 | 0.076 | undef | 0.244 |
|---|
| PFKFB1 |   | 1 | 1195 | 1323 | 1347 | -0.212 | -0.035 | 0.148 | -0.188 | 0.053 |
|---|
| BMPR2 |   | 5 | 360 | 1323 | 1292 | -0.132 | 0.081 | -0.029 | 0.356 | -0.021 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| CNBP |   | 1 | 1195 | 1323 | 1347 | -0.090 | -0.108 | 0.198 | undef | -0.057 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| NACAD |   | 3 | 557 | 1323 | 1309 | -0.159 | 0.096 | 0.093 | 0.155 | -0.078 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| TOMM70A |   | 1 | 1195 | 1323 | 1347 | -0.011 | 0.117 | -0.167 | 0.173 | 0.199 |
|---|
| GREM1 |   | 9 | 196 | 525 | 508 | -0.073 | 0.200 | 0.028 | 0.225 | 0.261 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| LDB1 |   | 2 | 743 | 1323 | 1313 | 0.138 | -0.079 | -0.010 | -0.051 | -0.100 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| SUPT5H |   | 6 | 301 | 1323 | 1281 | -0.085 | 0.007 | -0.298 | 0.002 | -0.068 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| IMP3 |   | 4 | 440 | 366 | 381 | 0.042 | -0.132 | 0.113 | -0.304 | -0.196 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| TPR |   | 11 | 148 | 727 | 701 | -0.130 | 0.120 | -0.036 | -0.085 | 0.156 |
|---|
| LMX1A |   | 1 | 1195 | 1323 | 1347 | undef | 0.306 | undef | undef | undef |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| POLR1C |   | 3 | 557 | 1323 | 1309 | 0.176 | -0.210 | 0.181 | -0.126 | 0.038 |
|---|
| YWHAH |   | 2 | 743 | 1323 | 1313 | -0.082 | -0.046 | 0.001 | -0.094 | -0.008 |
|---|
| SNCAIP |   | 10 | 167 | 366 | 362 | -0.091 | 0.013 | 0.092 | 0.136 | 0.059 |
|---|
| TRMT61A |   | 1 | 1195 | 1323 | 1347 | 0.118 | -0.030 | 0.122 | undef | -0.128 |
|---|
GO Enrichment output for subnetwork 6821 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 6.558E-12 | 1.602E-08 |
|---|
| protein sumoylation | GO:0016925 |  | 1.247E-07 | 1.523E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 7.95E-07 | 6.474E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 2.443E-06 | 1.492E-03 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 2.443E-06 | 1.193E-03 |
|---|
| pancreas development | GO:0031016 |  | 2.767E-06 | 1.127E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 4.259E-06 | 1.486E-03 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 6.789E-06 | 2.073E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.789E-06 | 1.843E-03 |
|---|
| lung alveolus development | GO:0048286 |  | 1.444E-05 | 3.528E-03 |
|---|
| motor axon guidance | GO:0008045 |  | 1.444E-05 | 3.207E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 4.726E-12 | 1.137E-08 |
|---|
| protein sumoylation | GO:0016925 |  | 1.693E-07 | 2.037E-04 |
|---|
| epithelial to mesenchymal transition | GO:0001837 |  | 1.077E-06 | 8.641E-04 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 1.544E-06 | 9.288E-04 |
|---|
| myoblast cell fate commitment | GO:0048625 |  | 3.076E-06 | 1.48E-03 |
|---|
| pancreas development | GO:0031016 |  | 3.745E-06 | 1.502E-03 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.361E-06 | 1.843E-03 |
|---|
| positive regulation of insulin secretion | GO:0032024 |  | 8.544E-06 | 2.57E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 8.544E-06 | 2.284E-03 |
|---|
| cell cycle arrest | GO:0007050 |  | 1.795E-05 | 4.319E-03 |
|---|
| lung alveolus development | GO:0048286 |  | 1.816E-05 | 3.973E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.096E-07 | 2.52E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 3.254E-07 | 3.742E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.048E-06 | 3.87E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.4E-05 | 8.052E-03 |
|---|
| lung alveolus development | GO:0048286 |  | 2.091E-05 | 9.618E-03 |
|---|
| regulation of homeostatic process | GO:0032844 |  | 4.07E-05 | 0.0155999 |
|---|
| positive regulation of bone mineralization | GO:0030501 |  | 6.987E-05 | 0.02295597 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.171E-05 | 0.02061706 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 8.851E-05 | 0.02261809 |
|---|
| positive regulation of biomineral formation | GO:0070169 |  | 8.851E-05 | 0.02035628 |
|---|
| regulation of erythrocyte differentiation | GO:0045646 |  | 8.851E-05 | 0.01850571 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cell migration | GO:0030334 |  | 2.837E-06 | 5.228E-03 |
|---|
| protein sumoylation | GO:0016925 |  | 3.598E-06 | 3.316E-03 |
|---|
| regulation of locomotion | GO:0040012 |  | 5.321E-06 | 3.269E-03 |
|---|
| regulation of cell motion | GO:0051270 |  | 5.321E-06 | 2.452E-03 |
|---|
| regulation of anatomical structure morphogenesis | GO:0022603 |  | 8.736E-06 | 3.22E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 1.135E-05 | 3.486E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.561E-05 | 4.111E-03 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 2.753E-05 | 6.342E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 2.909E-05 | 5.957E-03 |
|---|
| regulation of angiogenesis | GO:0045765 |  | 4.007E-05 | 7.384E-03 |
|---|
| lung development | GO:0030324 |  | 5.635E-05 | 9.441E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 1.096E-07 | 2.52E-04 |
|---|
| protein sumoylation | GO:0016925 |  | 3.254E-07 | 3.742E-04 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 5.048E-06 | 3.87E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.4E-05 | 8.052E-03 |
|---|
| lung alveolus development | GO:0048286 |  | 2.091E-05 | 9.618E-03 |
|---|
| regulation of homeostatic process | GO:0032844 |  | 4.07E-05 | 0.0155999 |
|---|
| positive regulation of bone mineralization | GO:0030501 |  | 6.987E-05 | 0.02295597 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 7.171E-05 | 0.02061706 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 8.851E-05 | 0.02261809 |
|---|
| positive regulation of biomineral formation | GO:0070169 |  | 8.851E-05 | 0.02035628 |
|---|
| regulation of erythrocyte differentiation | GO:0045646 |  | 8.851E-05 | 0.01850571 |
|---|



