Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6786
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7499 | 3.773e-03 | 6.270e-04 | 7.805e-01 | 1.846e-06 |
|---|
| Loi | 0.2309 | 7.941e-02 | 1.114e-02 | 4.261e-01 | 3.768e-04 |
|---|
| Schmidt | 0.6752 | 0.000e+00 | 0.000e+00 | 4.299e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.435e-03 | 0.000e+00 |
|---|
| Wang | 0.2634 | 2.441e-03 | 4.161e-02 | 3.859e-01 | 3.919e-05 |
|---|
Expression data for subnetwork 6786 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
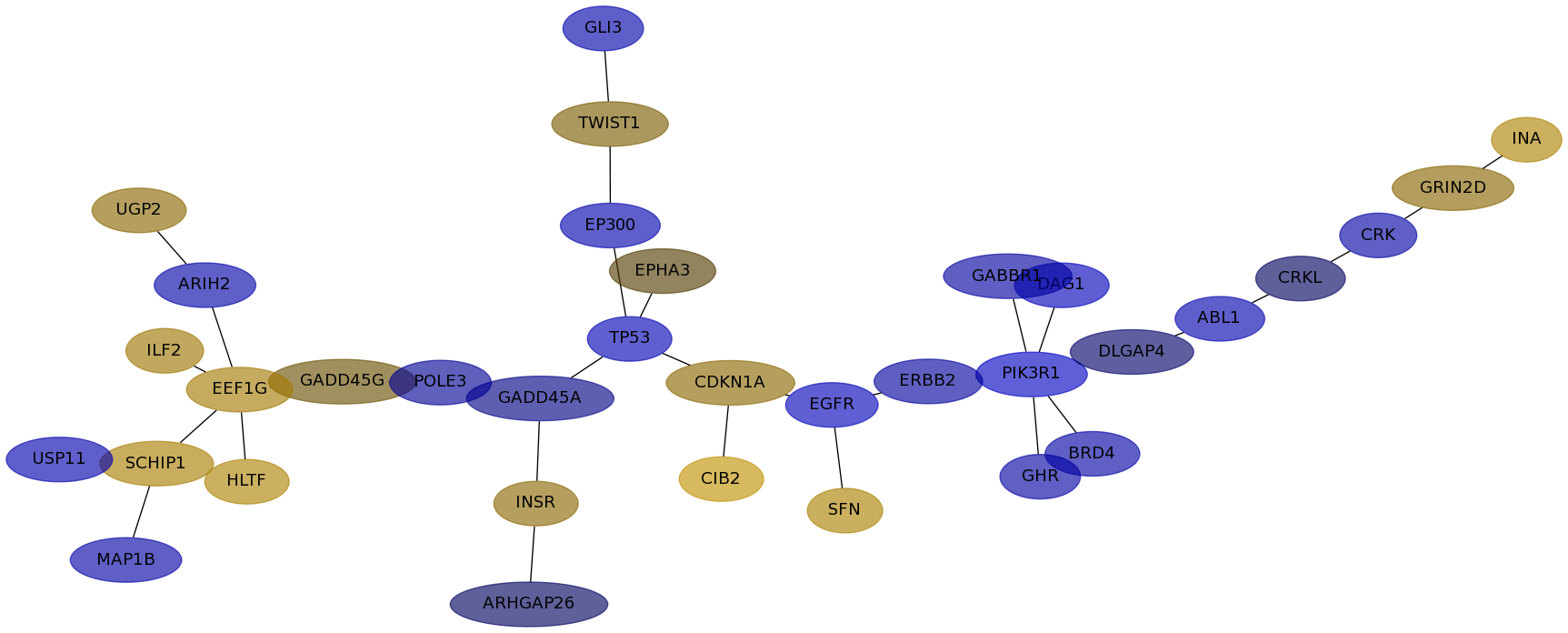
Score for each gene in subnetwork 6786 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| EPHA3 |   | 1 | 1195 | 1644 | 1661 | 0.016 | -0.044 | 0.188 | 0.116 | 0.115 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| UGP2 |   | 1 | 1195 | 1644 | 1661 | 0.064 | 0.008 | 0.097 | 0.196 | -0.139 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| GABBR1 |   | 5 | 360 | 780 | 767 | -0.107 | 0.164 | 0.081 | 0.074 | 0.131 |
|---|
| DLGAP4 |   | 7 | 256 | 727 | 714 | -0.029 | 0.180 | -0.201 | 0.135 | -0.043 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| HLTF |   | 2 | 743 | 1644 | 1630 | 0.145 | 0.070 | 0.114 | 0.209 | -0.006 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| EEF1G |   | 8 | 222 | 882 | 869 | 0.116 | -0.224 | 0.131 | 0.086 | 0.082 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| CRKL |   | 6 | 301 | 780 | 764 | -0.023 | 0.056 | 0.271 | -0.025 | -0.008 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| SCHIP1 |   | 10 | 167 | 478 | 459 | 0.130 | 0.136 | 0.105 | -0.022 | 0.198 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| POLE3 |   | 17 | 95 | 141 | 138 | -0.083 | -0.033 | 0.231 | 0.096 | 0.011 |
|---|
| ILF2 |   | 2 | 743 | 1558 | 1545 | 0.099 | -0.123 | -0.068 | 0.116 | 0.004 |
|---|
| BRD4 |   | 11 | 148 | 1047 | 1003 | -0.119 | 0.095 | -0.075 | 0.081 | -0.044 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| DAG1 |   | 2 | 743 | 1644 | 1630 | -0.197 | 0.258 | 0.049 | -0.076 | 0.039 |
|---|
| INA |   | 1 | 1195 | 1644 | 1661 | 0.144 | 0.025 | undef | undef | undef |
|---|
| ARIH2 |   | 5 | 360 | 236 | 242 | -0.131 | 0.196 | -0.071 | -0.127 | -0.016 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| CRK |   | 1 | 1195 | 1644 | 1661 | -0.116 | 0.106 | 0.090 | -0.001 | 0.017 |
|---|
| GADD45A |   | 11 | 148 | 1351 | 1308 | -0.057 | 0.076 | -0.099 | 0.109 | 0.093 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| USP11 |   | 7 | 256 | 478 | 462 | -0.144 | 0.106 | 0.121 | 0.218 | -0.044 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6786 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.237E-07 | 3.022E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.136E-07 | 5.052E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 5.326E-07 | 4.337E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 6.281E-07 | 3.836E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 8.568E-07 | 4.187E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 9.619E-07 | 3.916E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.083E-06 | 3.779E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.164E-06 | 3.554E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.44E-06 | 3.909E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.536E-06 | 3.753E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 4.766E-06 | 1.059E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.885E-07 | 4.534E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 6.287E-07 | 7.563E-04 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 7.487E-07 | 6.005E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 8.829E-07 | 5.311E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 1.204E-06 | 5.794E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.246E-06 | 4.996E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.418E-06 | 4.874E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.527E-06 | 4.593E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.865E-06 | 4.986E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.032E-06 | 4.888E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 7.2E-06 | 1.575E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.087E-07 | 7.1E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.069E-06 | 1.229E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.288E-06 | 9.874E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.288E-06 | 7.405E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 1.83E-06 | 8.418E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.874E-06 | 7.184E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.045E-06 | 6.72E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.466E-06 | 7.089E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.466E-06 | 6.301E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.633E-06 | 6.057E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.359E-05 | 2.841E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 7.673E-07 | 1.414E-03 |
|---|
| heart development | GO:0007507 |  | 8.219E-07 | 7.574E-04 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 8.797E-07 | 5.405E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.067E-06 | 4.916E-04 |
|---|
| positive regulation of growth | GO:0045927 |  | 1.583E-06 | 5.836E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.98E-06 | 9.153E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.612E-06 | 9.511E-04 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 4.778E-06 | 1.101E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.352E-06 | 1.301E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 8.711E-06 | 1.605E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.066E-06 | 1.519E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.087E-07 | 7.1E-04 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.069E-06 | 1.229E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 1.288E-06 | 9.874E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.288E-06 | 7.405E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 1.83E-06 | 8.418E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.874E-06 | 7.184E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.045E-06 | 6.72E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.466E-06 | 7.089E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.466E-06 | 6.301E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.633E-06 | 6.057E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.359E-05 | 2.841E-03 |
|---|



