Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6746
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7507 | 3.722e-03 | 6.160e-04 | 7.786e-01 | 1.785e-06 |
|---|
| Loi | 0.2310 | 7.916e-02 | 1.107e-02 | 4.255e-01 | 3.728e-04 |
|---|
| Schmidt | 0.6748 | 0.000e+00 | 0.000e+00 | 4.345e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.437e-03 | 0.000e+00 |
|---|
| Wang | 0.2632 | 2.472e-03 | 4.190e-02 | 3.873e-01 | 4.012e-05 |
|---|
Expression data for subnetwork 6746 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
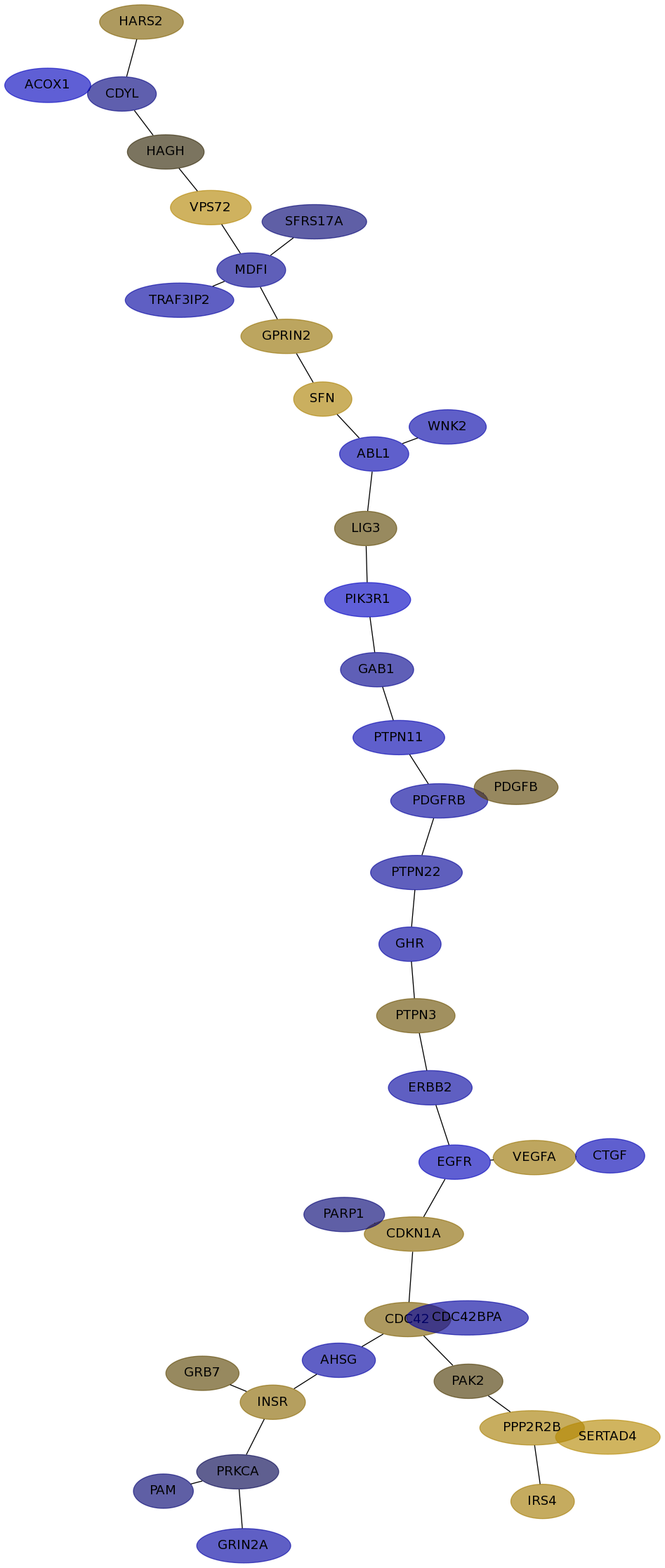
Score for each gene in subnetwork 6746 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| SFRS17A |   | 3 | 557 | 913 | 908 | -0.036 | 0.169 | -0.225 | -0.176 | -0.078 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PTPN3 |   | 11 | 148 | 658 | 635 | 0.029 | 0.060 | -0.067 | 0.225 | -0.034 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| LIG3 |   | 1 | 1195 | 1268 | 1293 | 0.022 | -0.203 | -0.115 | 0.176 | undef |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| PDGFB |   | 7 | 256 | 1165 | 1135 | 0.020 | 0.046 | 0.085 | -0.049 | 0.338 |
|---|
| ABL1 |   | 13 | 124 | 727 | 699 | -0.152 | 0.200 | 0.108 | undef | -0.059 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| VPS72 |   | 6 | 301 | 318 | 330 | 0.164 | -0.182 | 0.095 | 0.151 | 0.080 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| HARS2 |   | 3 | 557 | 1268 | 1255 | 0.049 | 0.360 | -0.168 | undef | -0.199 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| GRIN2A |   | 12 | 140 | 179 | 192 | -0.113 | -0.068 | 0.162 | 0.125 | undef |
|---|
| WNK2 |   | 7 | 256 | 1 | 47 | -0.127 | 0.029 | undef | 0.201 | undef |
|---|
| PDGFRB |   | 12 | 140 | 141 | 142 | -0.094 | 0.224 | 0.127 | 0.153 | 0.189 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| SERTAD4 |   | 8 | 222 | 1268 | 1225 | 0.173 | 0.169 | undef | -0.050 | undef |
|---|
| CDYL |   | 2 | 743 | 1268 | 1259 | -0.050 | 0.116 | -0.203 | undef | 0.206 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| PTPN22 |   | 5 | 360 | 957 | 938 | -0.086 | -0.137 | -0.292 | 0.148 | -0.151 |
|---|
| CDC42BPA |   | 6 | 301 | 1268 | 1236 | -0.102 | 0.098 | 0.059 | 0.368 | 0.260 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| TRAF3IP2 |   | 1 | 1195 | 1268 | 1293 | -0.105 | 0.129 | 0.042 | undef | 0.161 |
|---|
| HAGH |   | 2 | 743 | 1247 | 1238 | 0.007 | -0.155 | -0.128 | 0.202 | 0.033 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| AHSG |   | 5 | 360 | 412 | 416 | -0.113 | -0.054 | 0.000 | 0.132 | undef |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| ACOX1 |   | 9 | 196 | 1 | 44 | -0.209 | 0.122 | -0.104 | 0.252 | -0.094 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| PAM |   | 10 | 167 | 638 | 621 | -0.035 | 0.307 | 0.200 | 0.002 | 0.104 |
|---|
| GAB1 |   | 5 | 360 | 1102 | 1076 | -0.069 | 0.084 | -0.117 | 0.116 | -0.212 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6746 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.949E-13 | 4.762E-10 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 3.779E-12 | 4.616E-09 |
|---|
| indolalkylamine metabolic process | GO:0006586 |  | 2.458E-10 | 2.002E-07 |
|---|
| melatonin biosynthetic process | GO:0030187 |  | 3.45E-10 | 2.107E-07 |
|---|
| nuclear migration | GO:0007097 |  | 1.033E-09 | 5.045E-07 |
|---|
| indolalkylamine biosynthetic process | GO:0046219 |  | 1.033E-09 | 4.204E-07 |
|---|
| activation of MAPK activity | GO:0000187 |  | 1.126E-09 | 3.93E-07 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.39E-09 | 4.246E-07 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.404E-09 | 6.525E-07 |
|---|
| organ growth | GO:0035265 |  | 2.404E-09 | 5.873E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 2.404E-09 | 5.339E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 3.137E-13 | 7.549E-10 |
|---|
| indolalkylamine metabolic process | GO:0006586 |  | 1.146E-10 | 1.379E-07 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.797E-10 | 1.441E-07 |
|---|
| indolalkylamine biosynthetic process | GO:0046219 |  | 4.249E-10 | 2.556E-07 |
|---|
| nuclear migration | GO:0007097 |  | 1.272E-09 | 6.12E-07 |
|---|
| activation of MAPK activity | GO:0000187 |  | 1.591E-09 | 6.381E-07 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 1.591E-09 | 5.469E-07 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 2.961E-09 | 8.904E-07 |
|---|
| organ growth | GO:0035265 |  | 2.961E-09 | 7.915E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 2.961E-09 | 7.123E-07 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 2.961E-09 | 6.476E-07 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.553E-12 | 3.572E-09 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 6.005E-10 | 6.906E-07 |
|---|
| nuclear migration | GO:0007097 |  | 1.245E-09 | 9.544E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.245E-09 | 7.158E-07 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.723E-09 | 1.713E-06 |
|---|
| activation of MAPK activity | GO:0000187 |  | 3.882E-09 | 1.488E-06 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 6.216E-09 | 2.042E-06 |
|---|
| organ growth | GO:0035265 |  | 8.661E-09 | 2.49E-06 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 8.661E-09 | 2.213E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 8.661E-09 | 1.992E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 8.661E-09 | 1.811E-06 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 5.163E-12 | 9.515E-09 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 1.58E-11 | 1.456E-08 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 6.319E-11 | 3.882E-08 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 5.57E-10 | 2.566E-07 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 5.586E-10 | 2.059E-07 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.831E-09 | 8.695E-07 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 4.706E-09 | 1.239E-06 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.994E-08 | 4.593E-06 |
|---|
| regulation of protein amino acid phosphorylation | GO:0001932 |  | 7.579E-08 | 1.552E-05 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 9.64E-08 | 1.777E-05 |
|---|
| regulation of cell migration | GO:0030334 |  | 1.907E-07 | 3.194E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 1.553E-12 | 3.572E-09 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 6.005E-10 | 6.906E-07 |
|---|
| nuclear migration | GO:0007097 |  | 1.245E-09 | 9.544E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.245E-09 | 7.158E-07 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 3.723E-09 | 1.713E-06 |
|---|
| activation of MAPK activity | GO:0000187 |  | 3.882E-09 | 1.488E-06 |
|---|
| regulation of peptidyl-tyrosine phosphorylation | GO:0050730 |  | 6.216E-09 | 2.042E-06 |
|---|
| organ growth | GO:0035265 |  | 8.661E-09 | 2.49E-06 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 8.661E-09 | 2.213E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 8.661E-09 | 1.992E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 8.661E-09 | 1.811E-06 |
|---|



