Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6735
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7508 | 3.712e-03 | 6.140e-04 | 7.783e-01 | 1.774e-06 |
|---|
| Loi | 0.2310 | 7.917e-02 | 1.107e-02 | 4.255e-01 | 3.730e-04 |
|---|
| Schmidt | 0.6747 | 0.000e+00 | 0.000e+00 | 4.350e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.435e-03 | 0.000e+00 |
|---|
| Wang | 0.2630 | 2.488e-03 | 4.205e-02 | 3.880e-01 | 4.060e-05 |
|---|
Expression data for subnetwork 6735 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
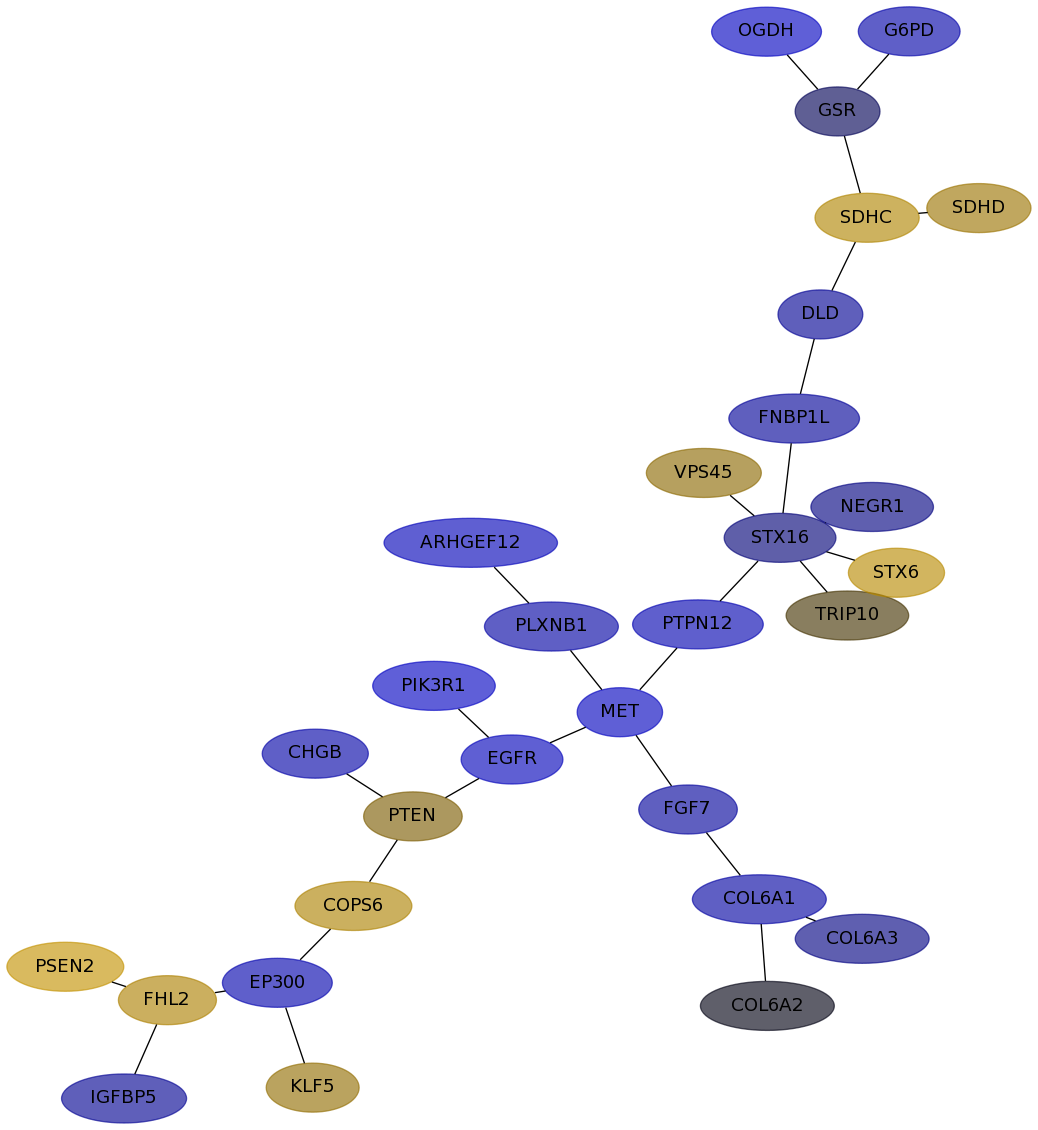
Score for each gene in subnetwork 6735 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| CHGB |   | 7 | 256 | 682 | 664 | -0.126 | -0.041 | 0.018 | 0.102 | -0.131 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| COL6A2 |   | 1 | 1195 | 1316 | 1336 | -0.002 | 0.115 | -0.071 | undef | 0.023 |
|---|
| SDHC |   | 3 | 557 | 1141 | 1129 | 0.158 | -0.033 | -0.077 | 0.199 | 0.110 |
|---|
| TRIP10 |   | 5 | 360 | 1034 | 1004 | 0.012 | 0.154 | 0.049 | undef | 0.144 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| VPS45 |   | 1 | 1195 | 1316 | 1336 | 0.066 | 0.029 | 0.000 | 0.183 | 0.067 |
|---|
| PTPN12 |   | 6 | 301 | 478 | 473 | -0.159 | 0.153 | 0.031 | 0.083 | 0.042 |
|---|
| ARHGEF12 |   | 2 | 743 | 1316 | 1312 | -0.190 | 0.167 | -0.018 | 0.206 | 0.126 |
|---|
| OGDH |   | 1 | 1195 | 1316 | 1336 | -0.229 | 0.138 | -0.289 | 0.132 | -0.151 |
|---|
| STX16 |   | 2 | 743 | 1034 | 1039 | -0.041 | 0.027 | 0.279 | 0.073 | 0.088 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| COL6A3 |   | 2 | 743 | 1077 | 1077 | -0.053 | 0.162 | 0.058 | 0.158 | 0.153 |
|---|
| PLXNB1 |   | 4 | 440 | 1047 | 1032 | -0.117 | 0.312 | 0.006 | -0.172 | -0.146 |
|---|
| NEGR1 |   | 1 | 1195 | 1316 | 1336 | -0.051 | 0.072 | undef | undef | undef |
|---|
| STX6 |   | 3 | 557 | 236 | 265 | 0.193 | 0.152 | 0.038 | 0.052 | 0.064 |
|---|
| SDHD |   | 1 | 1195 | 1316 | 1336 | 0.094 | 0.031 | 0.233 | -0.038 | -0.027 |
|---|
| GSR |   | 3 | 557 | 1222 | 1210 | -0.018 | -0.011 | -0.194 | 0.202 | -0.126 |
|---|
| PTEN |   | 27 | 50 | 141 | 135 | 0.045 | 0.055 | 0.017 | 0.183 | -0.041 |
|---|
| G6PD |   | 2 | 743 | 83 | 123 | -0.132 | -0.041 | -0.255 | 0.230 | 0.012 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| COL6A1 |   | 5 | 360 | 780 | 767 | -0.112 | 0.193 | -0.013 | undef | 0.039 |
|---|
| PSEN2 |   | 14 | 117 | 614 | 591 | 0.239 | -0.068 | -0.157 | 0.295 | 0.045 |
|---|
| FGF7 |   | 10 | 167 | 780 | 760 | -0.094 | 0.237 | 0.070 | 0.038 | 0.083 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| MET |   | 6 | 301 | 1047 | 1020 | -0.219 | 0.086 | 0.183 | -0.025 | 0.170 |
|---|
| COPS6 |   | 16 | 104 | 1 | 31 | 0.147 | -0.068 | 0.128 | 0.216 | 0.065 |
|---|
| FNBP1L |   | 3 | 557 | 1294 | 1276 | -0.088 | 0.204 | undef | 0.154 | undef |
|---|
| DLD |   | 1 | 1195 | 1316 | 1336 | -0.081 | 0.025 | -0.048 | 0.113 | -0.073 |
|---|
GO Enrichment output for subnetwork 6735 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 2.288E-10 | 5.591E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 6.851E-10 | 8.369E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.595E-09 | 1.299E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.184E-09 | 1.945E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 9.514E-09 | 4.649E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.492E-08 | 6.075E-06 |
|---|
| endothelial cell migration | GO:0043542 |  | 1.063E-07 | 3.71E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.063E-07 | 3.246E-05 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 1.063E-07 | 2.886E-05 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 1.461E-07 | 3.57E-05 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.724E-07 | 3.83E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| phospholipid dephosphorylation | GO:0046839 |  | 2.188E-10 | 5.263E-07 |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 6.55E-10 | 7.879E-07 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 1.525E-09 | 1.223E-06 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 3.044E-09 | 1.831E-06 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 9.097E-09 | 4.377E-06 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 1.427E-08 | 5.721E-06 |
|---|
| endothelial cell migration | GO:0043542 |  | 1.017E-07 | 3.494E-05 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.017E-07 | 3.058E-05 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 1.017E-07 | 2.718E-05 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 1.017E-07 | 2.446E-05 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 1.376E-07 | 3.01E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.634E-07 | 8.358E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 7.25E-07 | 8.338E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.266E-06 | 9.704E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 2.02E-06 | 1.162E-03 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 4.309E-06 | 1.982E-03 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 4.309E-06 | 1.652E-03 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.294E-05 | 4.253E-03 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 1.436E-05 | 4.128E-03 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 1.614E-05 | 4.125E-03 |
|---|
| endothelial cell migration | GO:0043542 |  | 1.982E-05 | 4.558E-03 |
|---|
| lipid modification | GO:0030258 |  | 2.4E-05 | 5.017E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| coenzyme metabolic process | GO:0006732 |  | 3.633E-06 | 6.696E-03 |
|---|
| coenzyme catabolic process | GO:0009109 |  | 9.134E-06 | 8.417E-03 |
|---|
| acetyl-CoA metabolic process | GO:0006084 |  | 1.16E-05 | 7.126E-03 |
|---|
| cofactor metabolic process | GO:0051186 |  | 1.185E-05 | 5.46E-03 |
|---|
| glutathione metabolic process | GO:0006749 |  | 1.447E-05 | 5.333E-03 |
|---|
| cofactor catabolic process | GO:0051187 |  | 1.777E-05 | 5.458E-03 |
|---|
| cellular respiration | GO:0045333 |  | 4.367E-05 | 0.01149679 |
|---|
| aerobic respiration | GO:0009060 |  | 4.823E-05 | 0.01111059 |
|---|
| regulation of cell migration | GO:0030334 |  | 7.952E-05 | 0.01628391 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 8.072E-05 | 0.01487609 |
|---|
| peptide metabolic process | GO:0006518 |  | 9.061E-05 | 0.01518102 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase B signaling cascade | GO:0051898 |  | 3.634E-07 | 8.358E-04 |
|---|
| negative regulation of focal adhesion formation | GO:0051895 |  | 7.25E-07 | 8.338E-04 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 1.266E-06 | 9.704E-04 |
|---|
| regulation of protein kinase B signaling cascade | GO:0051896 |  | 2.02E-06 | 1.162E-03 |
|---|
| negative regulation of cell-matrix adhesion | GO:0001953 |  | 4.309E-06 | 1.982E-03 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 4.309E-06 | 1.652E-03 |
|---|
| inositol metabolic process | GO:0006020 |  | 1.294E-05 | 4.253E-03 |
|---|
| negative regulation of cell migration | GO:0030336 |  | 1.436E-05 | 4.128E-03 |
|---|
| cardiac muscle tissue development | GO:0048738 |  | 1.614E-05 | 4.125E-03 |
|---|
| endothelial cell migration | GO:0043542 |  | 1.982E-05 | 4.558E-03 |
|---|
| lipid modification | GO:0030258 |  | 2.4E-05 | 5.017E-03 |
|---|



