Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6729
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7509 | 3.709e-03 | 6.130e-04 | 7.782e-01 | 1.769e-06 |
|---|
| Loi | 0.2311 | 7.902e-02 | 1.103e-02 | 4.252e-01 | 3.707e-04 |
|---|
| Schmidt | 0.6746 | 0.000e+00 | 0.000e+00 | 4.362e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7910 | 0.000e+00 | 0.000e+00 | 3.438e-03 | 0.000e+00 |
|---|
| Wang | 0.2630 | 2.497e-03 | 4.214e-02 | 3.884e-01 | 4.087e-05 |
|---|
Expression data for subnetwork 6729 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
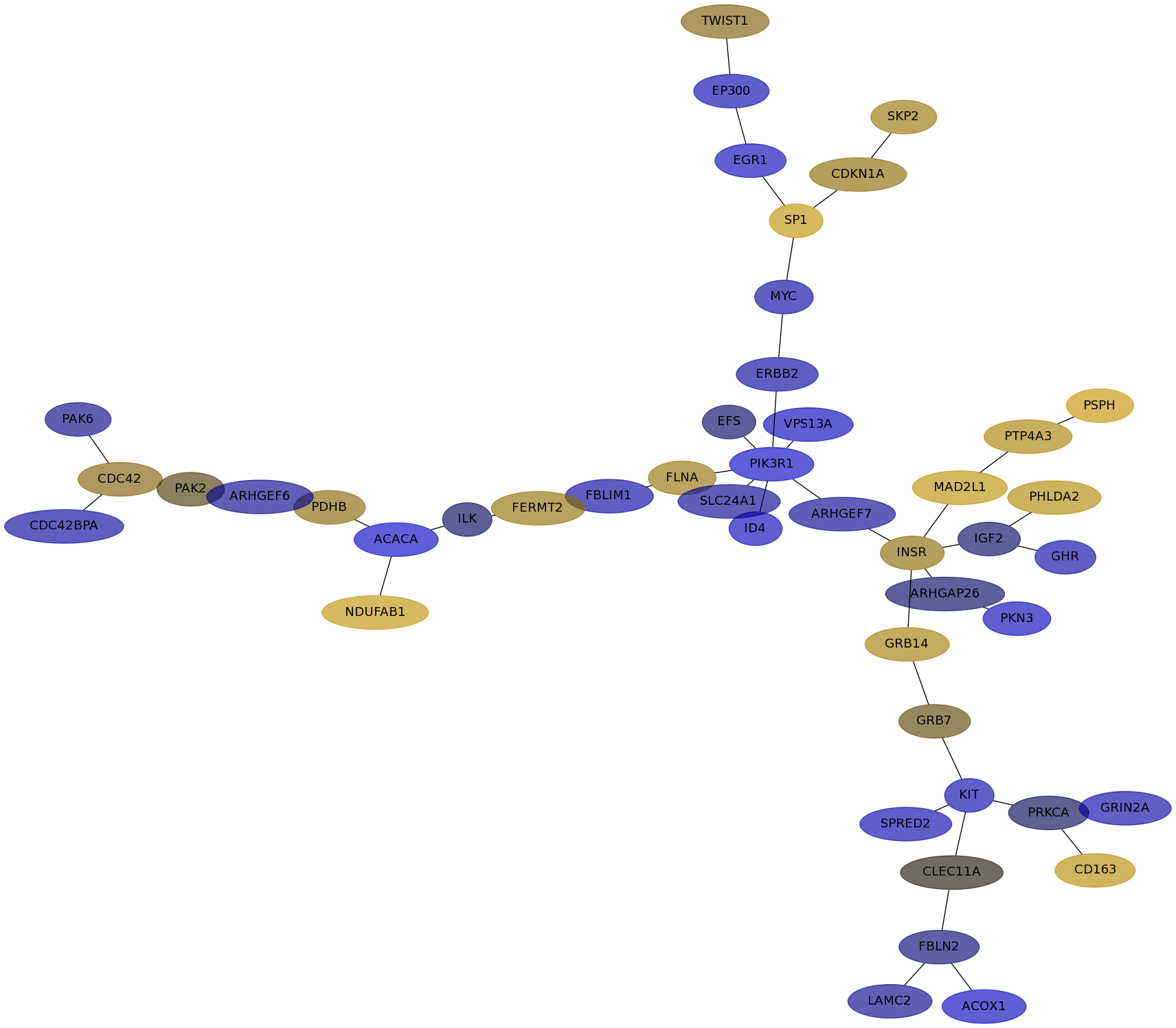
Score for each gene in subnetwork 6729 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| FBLIM1 |   | 1 | 1195 | 1413 | 1438 | -0.103 | 0.175 | undef | 0.119 | undef |
|---|
| SPRED2 |   | 2 | 743 | 1413 | 1410 | -0.151 | 0.224 | -0.047 | undef | 0.026 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| FBLN2 |   | 5 | 360 | 83 | 110 | -0.036 | 0.122 | 0.207 | 0.037 | -0.007 |
|---|
| GRB14 |   | 24 | 62 | 179 | 181 | 0.113 | -0.129 | 0.143 | -0.098 | 0.138 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| PTP4A3 |   | 9 | 196 | 412 | 411 | 0.137 | -0.196 | 0.085 | undef | -0.004 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| NDUFAB1 |   | 2 | 743 | 1413 | 1410 | 0.230 | -0.040 | 0.162 | -0.013 | -0.051 |
|---|
| EFS |   | 3 | 557 | 1323 | 1309 | -0.024 | 0.300 | -0.003 | 0.208 | -0.034 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| VPS13A |   | 10 | 167 | 236 | 234 | -0.203 | 0.105 | -0.046 | 0.120 | 0.062 |
|---|
| PHLDA2 |   | 2 | 743 | 957 | 958 | 0.158 | 0.164 | -0.007 | 0.258 | 0.210 |
|---|
| PAK6 |   | 1 | 1195 | 1413 | 1438 | -0.058 | -0.005 | 0.252 | -0.131 | 0.259 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| GRIN2A |   | 12 | 140 | 179 | 192 | -0.113 | -0.068 | 0.162 | 0.125 | undef |
|---|
| CD163 |   | 2 | 743 | 1413 | 1410 | 0.175 | -0.206 | -0.019 | -0.063 | 0.038 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| ACACA |   | 3 | 557 | 1413 | 1407 | -0.275 | -0.103 | 0.056 | undef | -0.080 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| CLEC11A |   | 7 | 256 | 692 | 673 | 0.004 | 0.353 | 0.005 | 0.081 | 0.272 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| FERMT2 |   | 2 | 743 | 1413 | 1410 | 0.077 | 0.151 | 0.219 | undef | 0.287 |
|---|
| CDC42BPA |   | 6 | 301 | 1268 | 1236 | -0.102 | 0.098 | 0.059 | 0.368 | 0.260 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| ILK |   | 4 | 440 | 1413 | 1390 | -0.020 | 0.195 | 0.076 | 0.130 | 0.086 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| SLC24A1 |   | 4 | 440 | 1323 | 1295 | -0.068 | 0.109 | 0.178 | -0.020 | 0.056 |
|---|
| PKN3 |   | 18 | 86 | 179 | 187 | -0.181 | 0.009 | undef | 0.239 | undef |
|---|
| ARHGEF6 |   | 10 | 167 | 525 | 507 | -0.068 | -0.120 | 0.166 | undef | -0.115 |
|---|
| ACOX1 |   | 9 | 196 | 1 | 44 | -0.209 | 0.122 | -0.104 | 0.252 | -0.094 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| MAD2L1 |   | 8 | 222 | 236 | 236 | 0.208 | -0.130 | 0.116 | 0.032 | 0.063 |
|---|
| LAMC2 |   | 4 | 440 | 179 | 209 | -0.063 | 0.307 | -0.060 | -0.148 | 0.055 |
|---|
| IGF2 |   | 14 | 117 | 570 | 553 | -0.022 | 0.172 | 0.145 | 0.087 | 0.116 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
GO Enrichment output for subnetwork 6729 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 2.318E-09 | 5.664E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.076E-08 | 1.314E-05 |
|---|
| nucleus localization | GO:0051647 |  | 5.03E-08 | 4.096E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.842E-07 | 1.125E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.009E-07 | 9.814E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.805E-07 | 1.142E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.861E-07 | 2.046E-04 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 8.84E-07 | 2.699E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 8.84E-07 | 2.399E-04 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 1.082E-06 | 2.642E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.543E-06 | 3.427E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 2.513E-09 | 6.045E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.166E-08 | 1.403E-05 |
|---|
| nucleus localization | GO:0051647 |  | 5.45E-08 | 4.371E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.722E-07 | 1.036E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.883E-07 | 9.063E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.657E-07 | 1.065E-04 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 4.716E-07 | 1.621E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.449E-07 | 1.94E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 9.406E-07 | 2.515E-04 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 9.61E-07 | 2.312E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 1.642E-06 | 3.591E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 2.719E-09 | 6.254E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.889E-08 | 2.172E-05 |
|---|
| nucleus localization | GO:0051647 |  | 6.749E-08 | 5.175E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.525E-07 | 2.027E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.916E-07 | 1.802E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.31E-07 | 2.035E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.141E-06 | 3.75E-04 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 1.237E-06 | 3.557E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.472E-06 | 3.761E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.274E-06 | 5.231E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.966E-06 | 8.293E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 8.235E-07 | 1.518E-03 |
|---|
| nucleus localization | GO:0051647 |  | 8.235E-07 | 7.588E-04 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 1.247E-06 | 7.662E-04 |
|---|
| peptidyl-tyrosine modification | GO:0018212 |  | 1.554E-06 | 7.161E-04 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 2.1E-06 | 7.742E-04 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.198E-06 | 6.752E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 5.591E-06 | 1.472E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 6.405E-06 | 1.475E-03 |
|---|
| negative regulation of cell cycle process | GO:0010948 |  | 6.833E-06 | 1.399E-03 |
|---|
| establishment of organelle localization | GO:0051656 |  | 8.519E-06 | 1.57E-03 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 9.763E-06 | 1.636E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| nuclear migration | GO:0007097 |  | 2.719E-09 | 6.254E-06 |
|---|
| establishment of nucleus localization | GO:0040023 |  | 1.889E-08 | 2.172E-05 |
|---|
| nucleus localization | GO:0051647 |  | 6.749E-08 | 5.175E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.525E-07 | 2.027E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.916E-07 | 1.802E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 5.31E-07 | 2.035E-04 |
|---|
| positive regulation of pseudopodium assembly | GO:0031274 |  | 1.141E-06 | 3.75E-04 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 1.237E-06 | 3.557E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.472E-06 | 3.761E-04 |
|---|
| macrophage differentiation | GO:0030225 |  | 2.274E-06 | 5.231E-04 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.966E-06 | 8.293E-04 |
|---|



