Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6707
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7513 | 3.677e-03 | 6.060e-04 | 7.770e-01 | 1.731e-06 |
|---|
| Loi | 0.2312 | 7.892e-02 | 1.101e-02 | 4.249e-01 | 3.691e-04 |
|---|
| Schmidt | 0.6745 | 0.000e+00 | 0.000e+00 | 4.376e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7909 | 0.000e+00 | 0.000e+00 | 3.453e-03 | 0.000e+00 |
|---|
| Wang | 0.2628 | 2.515e-03 | 4.231e-02 | 3.892e-01 | 4.141e-05 |
|---|
Expression data for subnetwork 6707 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
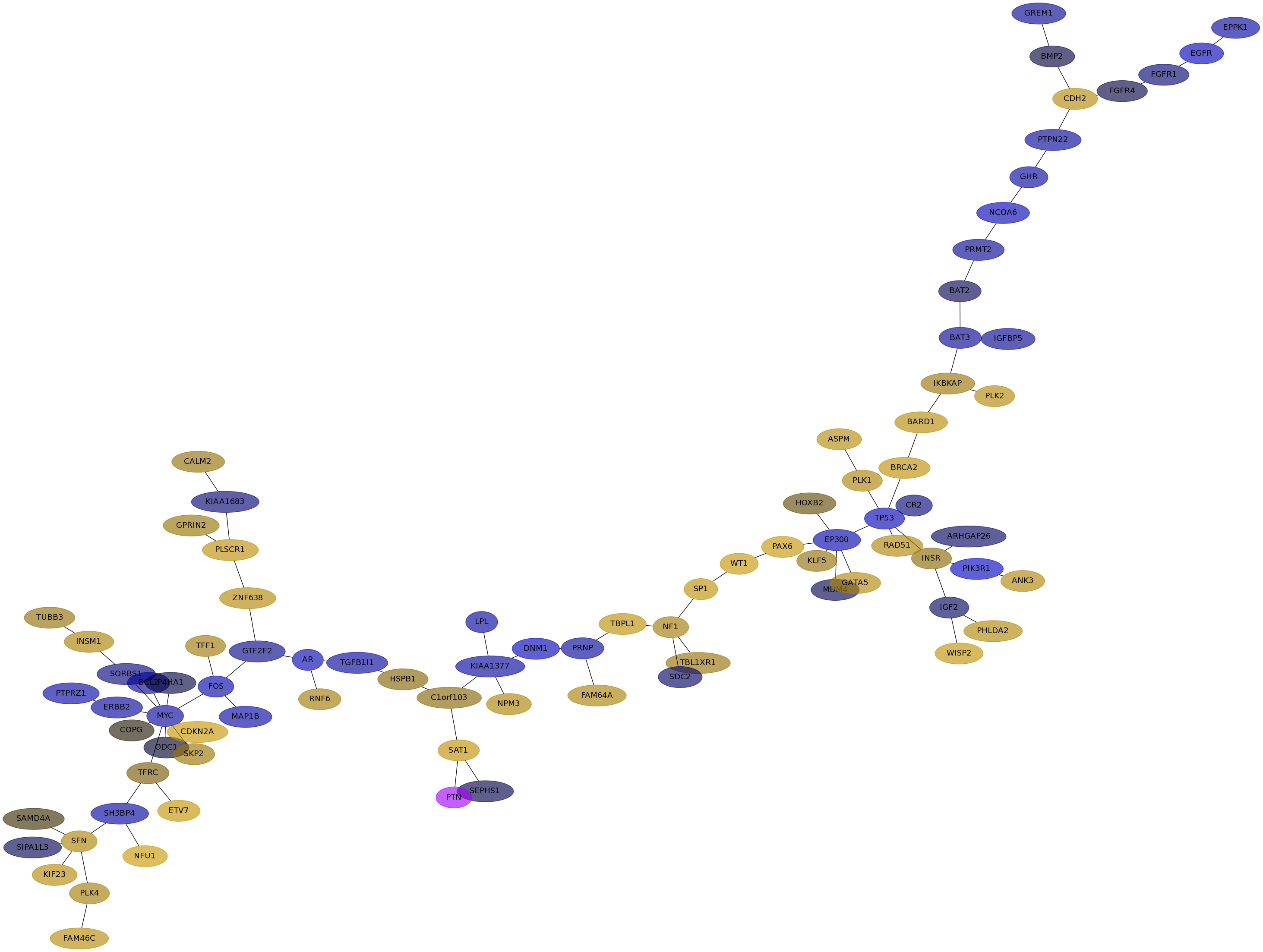
Score for each gene in subnetwork 6707 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NF1 |   | 9 | 196 | 552 | 541 | 0.110 | -0.052 | -0.032 | 0.090 | -0.094 |
|---|
| ANK3 |   | 6 | 301 | 83 | 105 | 0.153 | 0.034 | 0.001 | 0.094 | -0.038 |
|---|
| MDM4 |   | 9 | 196 | 141 | 147 | -0.016 | 0.004 | 0.164 | 0.107 | 0.105 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| FAM46C |   | 2 | 743 | 842 | 854 | 0.183 | -0.142 | -0.141 | -0.090 | -0.127 |
|---|
| PRNP |   | 2 | 743 | 957 | 958 | -0.090 | 0.168 | 0.020 | -0.222 | 0.048 |
|---|
| CALM2 |   | 2 | 743 | 957 | 958 | 0.076 | 0.056 | 0.104 | -0.004 | 0.086 |
|---|
| NCOA6 |   | 3 | 557 | 957 | 949 | -0.179 | -0.123 | -0.167 | 0.103 | 0.133 |
|---|
| PLK4 |   | 2 | 743 | 957 | 958 | 0.114 | -0.154 | 0.260 | 0.136 | 0.027 |
|---|
| NFU1 |   | 1 | 1195 | 957 | 992 | 0.280 | 0.023 | 0.085 | 0.107 | -0.257 |
|---|
| BAT3 |   | 4 | 440 | 236 | 262 | -0.078 | 0.072 | -0.057 | 0.110 | 0.069 |
|---|
| SEPHS1 |   | 3 | 557 | 957 | 949 | -0.014 | -0.311 | 0.129 | -0.023 | 0.033 |
|---|
| ZNF638 |   | 5 | 360 | 806 | 784 | 0.164 | 0.088 | 0.020 | 0.077 | -0.045 |
|---|
| PLK1 |   | 11 | 148 | 236 | 232 | 0.142 | -0.143 | 0.094 | -0.192 | -0.163 |
|---|
| NPM3 |   | 1 | 1195 | 957 | 992 | 0.142 | -0.110 | 0.317 | 0.042 | 0.228 |
|---|
| SAT1 |   | 6 | 301 | 236 | 239 | 0.219 | 0.010 | -0.066 | -0.119 | 0.063 |
|---|
| CR2 |   | 5 | 360 | 957 | 938 | -0.042 | -0.089 | 0.101 | 0.018 | 0.015 |
|---|
| PTPN22 |   | 5 | 360 | 957 | 938 | -0.086 | -0.137 | -0.292 | 0.148 | -0.151 |
|---|
| ODC1 |   | 4 | 440 | 141 | 165 | -0.006 | -0.099 | 0.209 | -0.207 | -0.107 |
|---|
| FGFR1 |   | 26 | 52 | 638 | 615 | -0.036 | 0.200 | 0.087 | undef | -0.015 |
|---|
| KIAA1683 |   | 1 | 1195 | 957 | 992 | -0.039 | 0.047 | undef | undef | undef |
|---|
| RNF6 |   | 4 | 440 | 590 | 585 | 0.109 | -0.128 | 0.002 | 0.041 | -0.018 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| PLK2 |   | 1 | 1195 | 957 | 992 | 0.165 | 0.258 | 0.089 | -0.127 | -0.173 |
|---|
| P4HA1 |   | 2 | 743 | 957 | 958 | -0.011 | 0.089 | -0.020 | undef | 0.169 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| GATA5 |   | 8 | 222 | 366 | 373 | 0.161 | -0.031 | undef | undef | undef |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| COPG |   | 5 | 360 | 570 | 562 | 0.004 | 0.080 | -0.158 | undef | 0.280 |
|---|
| SDC2 |   | 4 | 440 | 590 | 585 | -0.027 | 0.068 | 0.033 | undef | 0.215 |
|---|
| KIF23 |   | 4 | 440 | 957 | 942 | 0.168 | -0.063 | 0.162 | undef | 0.033 |
|---|
| KIAA1377 |   | 18 | 86 | 141 | 136 | -0.095 | 0.116 | undef | 0.198 | undef |
|---|
| IKBKAP |   | 10 | 167 | 412 | 408 | 0.089 | 0.184 | 0.142 | 0.105 | -0.017 |
|---|
| PHLDA2 |   | 2 | 743 | 957 | 958 | 0.158 | 0.164 | -0.007 | 0.258 | 0.210 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| TFRC |   | 5 | 360 | 957 | 938 | 0.038 | -0.066 | 0.076 | -0.108 | -0.014 |
|---|
| WISP2 |   | 3 | 557 | 957 | 949 | 0.232 | -0.010 | 0.088 | 0.047 | 0.204 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| SIPA1L3 |   | 3 | 557 | 648 | 647 | -0.016 | 0.039 | -0.062 | 0.136 | 0.049 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| GTF2F2 |   | 4 | 440 | 957 | 942 | -0.059 | -0.085 | 0.119 | -0.004 | -0.054 |
|---|
| PTPRZ1 |   | 9 | 196 | 570 | 554 | -0.114 | 0.183 | 0.233 | -0.316 | -0.095 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| INSM1 |   | 23 | 66 | 570 | 545 | 0.135 | -0.184 | -0.188 | 0.237 | -0.032 |
|---|
| ETV7 |   | 4 | 440 | 957 | 942 | 0.245 | -0.201 | -0.043 | -0.062 | -0.174 |
|---|
| C1orf103 |   | 4 | 440 | 412 | 419 | 0.057 | 0.029 | -0.173 | 0.127 | -0.159 |
|---|
| SAMD4A |   | 1 | 1195 | 957 | 992 | 0.009 | 0.134 | 0.189 | 0.021 | 0.187 |
|---|
| EPPK1 |   | 1 | 1195 | 957 | 992 | -0.102 | 0.186 | -0.032 | undef | 0.187 |
|---|
| TUBB3 |   | 7 | 256 | 764 | 756 | 0.078 | 0.130 | 0.073 | 0.207 | -0.079 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| FGFR4 |   | 21 | 73 | 179 | 183 | -0.011 | 0.188 | -0.089 | -0.018 | -0.049 |
|---|
| GREM1 |   | 9 | 196 | 525 | 508 | -0.073 | 0.200 | 0.028 | 0.225 | 0.261 |
|---|
| DNM1 |   | 4 | 440 | 236 | 262 | -0.203 | 0.095 | 0.163 | 0.001 | 0.082 |
|---|
| PLSCR1 |   | 10 | 167 | 412 | 408 | 0.202 | -0.159 | 0.091 | undef | -0.105 |
|---|
| TBL1XR1 |   | 3 | 557 | 552 | 558 | 0.084 | -0.064 | undef | undef | undef |
|---|
| BAT2 |   | 3 | 557 | 957 | 949 | -0.017 | 0.031 | 0.058 | undef | 0.016 |
|---|
| ASPM |   | 2 | 743 | 957 | 958 | 0.175 | 0.019 | undef | -0.001 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| PRMT2 |   | 5 | 360 | 957 | 938 | -0.081 | 0.046 | 0.089 | 0.235 | -0.128 |
|---|
| FOS |   | 42 | 27 | 1 | 17 | -0.137 | 0.129 | 0.185 | -0.144 | 0.234 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| TFF1 |   | 7 | 256 | 957 | 928 | 0.106 | 0.022 | 0.088 | -0.033 | -0.244 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| FAM64A |   | 1 | 1195 | 957 | 992 | 0.131 | -0.216 | 0.035 | -0.050 | 0.070 |
|---|
| RAD51 |   | 13 | 124 | 727 | 699 | 0.146 | -0.098 | 0.135 | -0.070 | 0.061 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| TBPL1 |   | 3 | 557 | 236 | 265 | 0.211 | -0.037 | 0.023 | 0.061 | 0.217 |
|---|
| SH3BP4 |   | 4 | 440 | 957 | 942 | -0.105 | 0.108 | -0.100 | 0.246 | -0.003 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| CDH2 |   | 10 | 167 | 727 | 702 | 0.169 | 0.109 | -0.016 | 0.123 | 0.155 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| IGF2 |   | 14 | 117 | 570 | 553 | -0.022 | 0.172 | 0.145 | 0.087 | 0.116 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6707 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 3.268E-09 | 7.984E-06 |
|---|
| urogenital system development | GO:0001655 |  | 2.328E-08 | 2.844E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.618E-08 | 2.132E-05 |
|---|
| metanephros development | GO:0001656 |  | 7.138E-08 | 4.359E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.413E-07 | 6.902E-05 |
|---|
| kidney development | GO:0001822 |  | 1.717E-07 | 6.993E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.825E-07 | 3.429E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 1.956E-06 | 5.974E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 1.956E-06 | 5.31E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 3.048E-06 | 7.446E-04 |
|---|
| stem cell division | GO:0017145 |  | 3.048E-06 | 6.769E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.09E-09 | 1.465E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 4.409E-08 | 5.304E-05 |
|---|
| urogenital system development | GO:0001655 |  | 5.184E-08 | 4.158E-05 |
|---|
| metanephros development | GO:0001656 |  | 1.32E-07 | 7.941E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.149E-07 | 1.034E-04 |
|---|
| kidney development | GO:0001822 |  | 3.465E-07 | 1.39E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.346E-06 | 4.625E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 2.965E-06 | 8.918E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 2.965E-06 | 7.927E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 4.179E-06 | 1.005E-03 |
|---|
| stem cell division | GO:0017145 |  | 4.179E-06 | 9.14E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 8.734E-09 | 2.009E-05 |
|---|
| urogenital system development | GO:0001655 |  | 7.678E-08 | 8.829E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.095E-07 | 8.394E-05 |
|---|
| metanephros development | GO:0001656 |  | 3.141E-07 | 1.806E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.483E-07 | 2.062E-04 |
|---|
| kidney development | GO:0001822 |  | 4.893E-07 | 1.876E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.16E-06 | 7.096E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 3.081E-06 | 8.859E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 6.143E-06 | 1.57E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 7.285E-06 | 1.676E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.274E-05 | 2.664E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.329E-07 | 6.135E-04 |
|---|
| urogenital system development | GO:0001655 |  | 6.845E-07 | 6.308E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 9.86E-07 | 6.057E-04 |
|---|
| positive regulation of growth | GO:0045927 |  | 1.167E-06 | 5.377E-04 |
|---|
| response to hormone stimulus | GO:0009725 |  | 1.808E-06 | 6.664E-04 |
|---|
| regulation of neurogenesis | GO:0050767 |  | 3.639E-06 | 1.118E-03 |
|---|
| T cell lineage commitment | GO:0002360 |  | 5.919E-06 | 1.558E-03 |
|---|
| negative regulation of neuroblast proliferation | GO:0007406 |  | 5.919E-06 | 1.364E-03 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 6.207E-06 | 1.271E-03 |
|---|
| kidney development | GO:0001822 |  | 6.584E-06 | 1.213E-03 |
|---|
| regulation of nervous system development | GO:0051960 |  | 7.146E-06 | 1.197E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 8.734E-09 | 2.009E-05 |
|---|
| urogenital system development | GO:0001655 |  | 7.678E-08 | 8.829E-05 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.095E-07 | 8.394E-05 |
|---|
| metanephros development | GO:0001656 |  | 3.141E-07 | 1.806E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.483E-07 | 2.062E-04 |
|---|
| kidney development | GO:0001822 |  | 4.893E-07 | 1.876E-04 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 2.16E-06 | 7.096E-04 |
|---|
| response to gamma radiation | GO:0010332 |  | 3.081E-06 | 8.859E-04 |
|---|
| branching involved in ureteric bud morphogenesis | GO:0001658 |  | 6.143E-06 | 1.57E-03 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 7.285E-06 | 1.676E-03 |
|---|
| response to peptide hormone stimulus | GO:0043434 |  | 1.274E-05 | 2.664E-03 |
|---|



