Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6685
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7518 | 3.644e-03 | 5.990e-04 | 7.757e-01 | 1.693e-06 |
|---|
| Loi | 0.2311 | 7.908e-02 | 1.105e-02 | 4.253e-01 | 3.716e-04 |
|---|
| Schmidt | 0.6743 | 0.000e+00 | 0.000e+00 | 4.401e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.458e-03 | 0.000e+00 |
|---|
| Wang | 0.2626 | 2.537e-03 | 4.251e-02 | 3.902e-01 | 4.208e-05 |
|---|
Expression data for subnetwork 6685 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
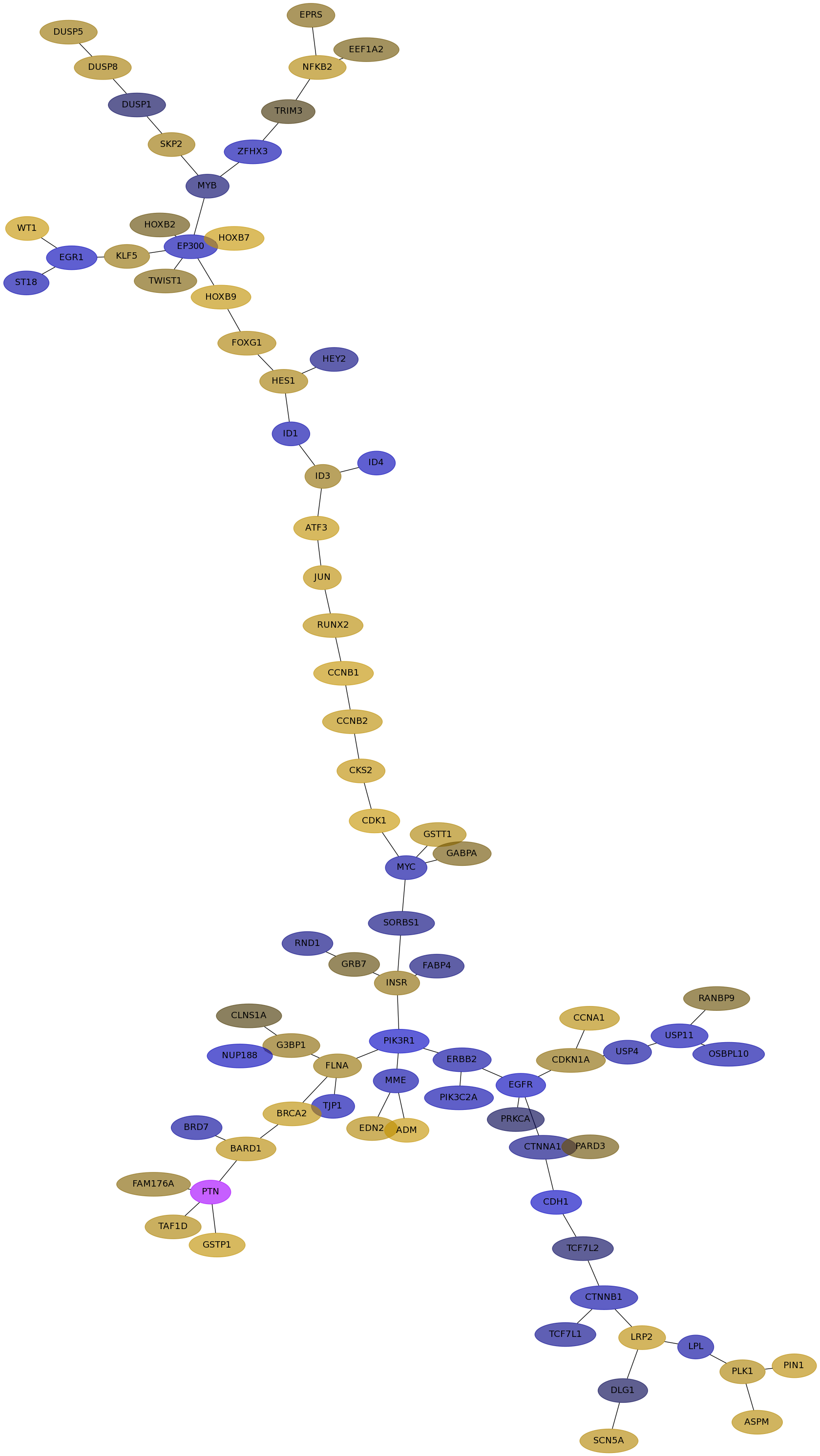
Score for each gene in subnetwork 6685 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ID1 |   | 3 | 557 | 806 | 792 | -0.128 | -0.030 | 0.124 | 0.069 | -0.142 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| NUP188 |   | 8 | 222 | 1116 | 1079 | -0.187 | 0.094 | 0.058 | 0.161 | -0.070 |
|---|
| HOXB7 |   | 6 | 301 | 1 | 52 | 0.267 | -0.139 | -0.083 | 0.085 | -0.051 |
|---|
| DUSP8 |   | 4 | 440 | 1 | 59 | 0.118 | 0.257 | 0.068 | 0.208 | -0.029 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| CCNB1 |   | 4 | 440 | 1116 | 1101 | 0.241 | -0.056 | 0.001 | undef | -0.043 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| BARD1 |   | 69 | 12 | 141 | 132 | 0.179 | -0.165 | 0.086 | 0.245 | -0.104 |
|---|
| HEY2 |   | 8 | 222 | 570 | 555 | -0.045 | -0.021 | 0.398 | -0.024 | -0.058 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| ZFHX3 |   | 1 | 1195 | 1116 | 1146 | -0.138 | -0.025 | -0.030 | 0.108 | -0.026 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| DLG1 |   | 6 | 301 | 457 | 452 | -0.016 | 0.173 | 0.244 | 0.017 | 0.123 |
|---|
| ST18 |   | 16 | 104 | 236 | 227 | -0.131 | 0.028 | -0.088 | 0.186 | 0.151 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| NFKB2 |   | 7 | 256 | 1116 | 1082 | 0.153 | -0.239 | -0.131 | undef | -0.065 |
|---|
| CKS2 |   | 9 | 196 | 928 | 903 | 0.214 | -0.220 | 0.089 | -0.003 | -0.056 |
|---|
| PLK1 |   | 11 | 148 | 236 | 232 | 0.142 | -0.143 | 0.094 | -0.192 | -0.163 |
|---|
| FAM176A |   | 2 | 743 | 1116 | 1121 | 0.054 | 0.217 | undef | undef | undef |
|---|
| GABPA |   | 23 | 66 | 696 | 665 | 0.033 | -0.183 | 0.066 | 0.141 | -0.161 |
|---|
| ATF3 |   | 17 | 95 | 179 | 189 | 0.225 | -0.021 | 0.000 | undef | -0.039 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| ADM |   | 10 | 167 | 682 | 653 | 0.255 | 0.187 | 0.091 | 0.169 | 0.212 |
|---|
| CLNS1A |   | 1 | 1195 | 1116 | 1146 | 0.013 | -0.095 | 0.092 | 0.150 | -0.081 |
|---|
| EDN2 |   | 3 | 557 | 1116 | 1107 | 0.153 | 0.062 | 0.132 | 0.196 | 0.287 |
|---|
| TCF7L1 |   | 2 | 743 | 1034 | 1039 | -0.062 | -0.031 | 0.211 | undef | -0.108 |
|---|
| BRCA2 |   | 41 | 28 | 236 | 222 | 0.206 | -0.124 | -0.066 | 0.233 | -0.116 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| LPL |   | 13 | 124 | 236 | 230 | -0.102 | -0.062 | 0.292 | 0.087 | -0.105 |
|---|
| USP11 |   | 7 | 256 | 478 | 462 | -0.144 | 0.106 | 0.121 | 0.218 | -0.044 |
|---|
| G3BP1 |   | 7 | 256 | 1116 | 1082 | 0.060 | -0.077 | -0.017 | 0.155 | -0.004 |
|---|
| HOXB9 |   | 2 | 743 | 1055 | 1059 | 0.231 | -0.101 | undef | undef | undef |
|---|
| PTN |   | 24 | 62 | 236 | 225 | undef | 0.022 | 0.000 | 0.054 | -0.092 |
|---|
| PARD3 |   | 6 | 301 | 366 | 377 | 0.030 | -0.006 | -0.052 | 0.040 | -0.090 |
|---|
| FABP4 |   | 6 | 301 | 236 | 239 | -0.036 | -0.028 | 0.227 | 0.072 | 0.107 |
|---|
| SCN5A |   | 1 | 1195 | 1116 | 1146 | 0.166 | -0.192 | 0.162 | -0.172 | 0.015 |
|---|
| RUNX2 |   | 2 | 743 | 1116 | 1121 | 0.184 | -0.104 | -0.127 | 0.013 | undef |
|---|
| USP4 |   | 7 | 256 | 1 | 47 | -0.093 | 0.147 | 0.097 | -0.392 | 0.054 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| CTNNA1 |   | 2 | 743 | 1116 | 1121 | -0.049 | 0.218 | 0.010 | 0.115 | -0.098 |
|---|
| LRP2 |   | 3 | 557 | 1102 | 1098 | 0.193 | -0.042 | 0.001 | -0.138 | 0.125 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| TJP1 |   | 8 | 222 | 318 | 325 | -0.139 | 0.102 | 0.099 | 0.319 | 0.049 |
|---|
| FOXG1 |   | 6 | 301 | 727 | 716 | 0.136 | -0.156 | -0.126 | -0.013 | -0.068 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| DUSP5 |   | 1 | 1195 | 1116 | 1146 | 0.090 | 0.008 | -0.051 | 0.051 | -0.035 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| RND1 |   | 6 | 301 | 236 | 239 | -0.046 | 0.085 | 0.022 | 0.082 | 0.113 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| EEF1A2 |   | 8 | 222 | 457 | 450 | 0.032 | 0.220 | -0.119 | 0.018 | 0.102 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| TRIM3 |   | 2 | 743 | 1116 | 1121 | 0.010 | 0.009 | -0.118 | undef | 0.063 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| DUSP1 |   | 4 | 440 | 1 | 59 | -0.019 | 0.127 | 0.170 | -0.031 | 0.134 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| HES1 |   | 4 | 440 | 570 | 565 | 0.115 | 0.157 | 0.010 | 0.214 | -0.106 |
|---|
| TAF1D |   | 10 | 167 | 590 | 566 | 0.141 | 0.113 | 0.041 | undef | 0.059 |
|---|
| ASPM |   | 2 | 743 | 957 | 958 | 0.175 | 0.019 | undef | -0.001 | undef |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| OSBPL10 |   | 2 | 743 | 780 | 787 | -0.116 | 0.239 | 0.026 | 0.187 | 0.035 |
|---|
| RANBP9 |   | 2 | 743 | 991 | 999 | 0.028 | 0.186 | 0.063 | 0.205 | 0.116 |
|---|
| PIK3C2A |   | 1 | 1195 | 1116 | 1146 | -0.130 | 0.037 | 0.165 | 0.091 | 0.067 |
|---|
| TCF7L2 |   | 17 | 95 | 366 | 358 | -0.021 | 0.143 | 0.280 | undef | 0.101 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| EPRS |   | 2 | 743 | 1116 | 1121 | 0.043 | -0.045 | -0.125 | 0.152 | 0.120 |
|---|
| ID3 |   | 9 | 196 | 658 | 637 | 0.072 | -0.006 | 0.192 | -0.023 | 0.057 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| CCNB2 |   | 4 | 440 | 842 | 831 | 0.206 | -0.119 | 0.004 | -0.006 | 0.134 |
|---|
| MME |   | 5 | 360 | 1047 | 1021 | -0.121 | 0.020 | 0.088 | -0.002 | -0.130 |
|---|
GO Enrichment output for subnetwork 6685 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.936E-09 | 7.173E-06 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.516E-07 | 3.073E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.539E-07 | 2.068E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 3.641E-07 | 2.224E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 3.892E-07 | 1.902E-04 |
|---|
| negative regulation of neuron differentiation | GO:0045665 |  | 6.927E-07 | 2.821E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 1.089E-06 | 3.801E-04 |
|---|
| regulation of the force of heart contraction | GO:0002026 |  | 1.479E-06 | 4.515E-04 |
|---|
| neutrophil chemotaxis | GO:0030593 |  | 2.554E-06 | 6.934E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 3.295E-06 | 8.049E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 3.586E-06 | 7.964E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| Wnt receptor signaling pathway through beta-catenin | GO:0060070 |  | 2.268E-09 | 5.456E-06 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.561E-07 | 4.284E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 3.749E-07 | 3.006E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 4.79E-07 | 2.881E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 5.455E-07 | 2.625E-04 |
|---|
| negative regulation of neuron differentiation | GO:0045665 |  | 6.661E-07 | 2.671E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 1.432E-06 | 4.921E-04 |
|---|
| regulation of the force of heart contraction | GO:0002026 |  | 1.943E-06 | 5.843E-04 |
|---|
| neutrophil chemotaxis | GO:0030593 |  | 3.354E-06 | 8.967E-04 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 3.418E-06 | 8.224E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.096E-06 | 8.96E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.128E-07 | 1.409E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.957E-07 | 9.15E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 1.108E-06 | 8.498E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.241E-06 | 7.134E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 2.373E-06 | 1.091E-03 |
|---|
| regulation of the force of heart contraction | GO:0002026 |  | 4.475E-06 | 1.716E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.199E-06 | 1.708E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.762E-06 | 1.657E-03 |
|---|
| neutrophil chemotaxis | GO:0030593 |  | 5.931E-06 | 1.516E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 6.143E-06 | 1.413E-03 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 7.709E-06 | 1.612E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cell junction assembly | GO:0034329 |  | 6.937E-06 | 0.01278471 |
|---|
| positive regulation of transcription factor import into nucleus | GO:0042993 |  | 8.495E-06 | 7.828E-03 |
|---|
| blood circulation | GO:0008015 |  | 8.651E-06 | 5.315E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 1.388E-05 | 6.396E-03 |
|---|
| cell junction organization | GO:0034330 |  | 2.072E-05 | 7.638E-03 |
|---|
| cell-cell junction assembly | GO:0007043 |  | 2.353E-05 | 7.227E-03 |
|---|
| actin filament organization | GO:0007015 |  | 2.981E-05 | 7.85E-03 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 3.51E-05 | 8.086E-03 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 3.523E-05 | 7.213E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 4.832E-05 | 8.905E-03 |
|---|
| negative regulation of neuron differentiation | GO:0045665 |  | 4.987E-05 | 8.356E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 6.128E-07 | 1.409E-03 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.957E-07 | 9.15E-04 |
|---|
| chondrocyte differentiation | GO:0002062 |  | 1.108E-06 | 8.498E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 1.241E-06 | 7.134E-04 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 2.373E-06 | 1.091E-03 |
|---|
| regulation of the force of heart contraction | GO:0002026 |  | 4.475E-06 | 1.716E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.199E-06 | 1.708E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.762E-06 | 1.657E-03 |
|---|
| neutrophil chemotaxis | GO:0030593 |  | 5.931E-06 | 1.516E-03 |
|---|
| establishment or maintenance of cell polarity | GO:0007163 |  | 6.143E-06 | 1.413E-03 |
|---|
| inactivation of MAPK activity | GO:0000188 |  | 7.709E-06 | 1.612E-03 |
|---|



