Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6666
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7521 | 3.628e-03 | 5.960e-04 | 7.751e-01 | 1.676e-06 |
|---|
| Loi | 0.2310 | 7.917e-02 | 1.107e-02 | 4.255e-01 | 3.730e-04 |
|---|
| Schmidt | 0.6740 | 0.000e+00 | 0.000e+00 | 4.428e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.472e-03 | 0.000e+00 |
|---|
| Wang | 0.2627 | 2.533e-03 | 4.247e-02 | 3.900e-01 | 4.196e-05 |
|---|
Expression data for subnetwork 6666 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
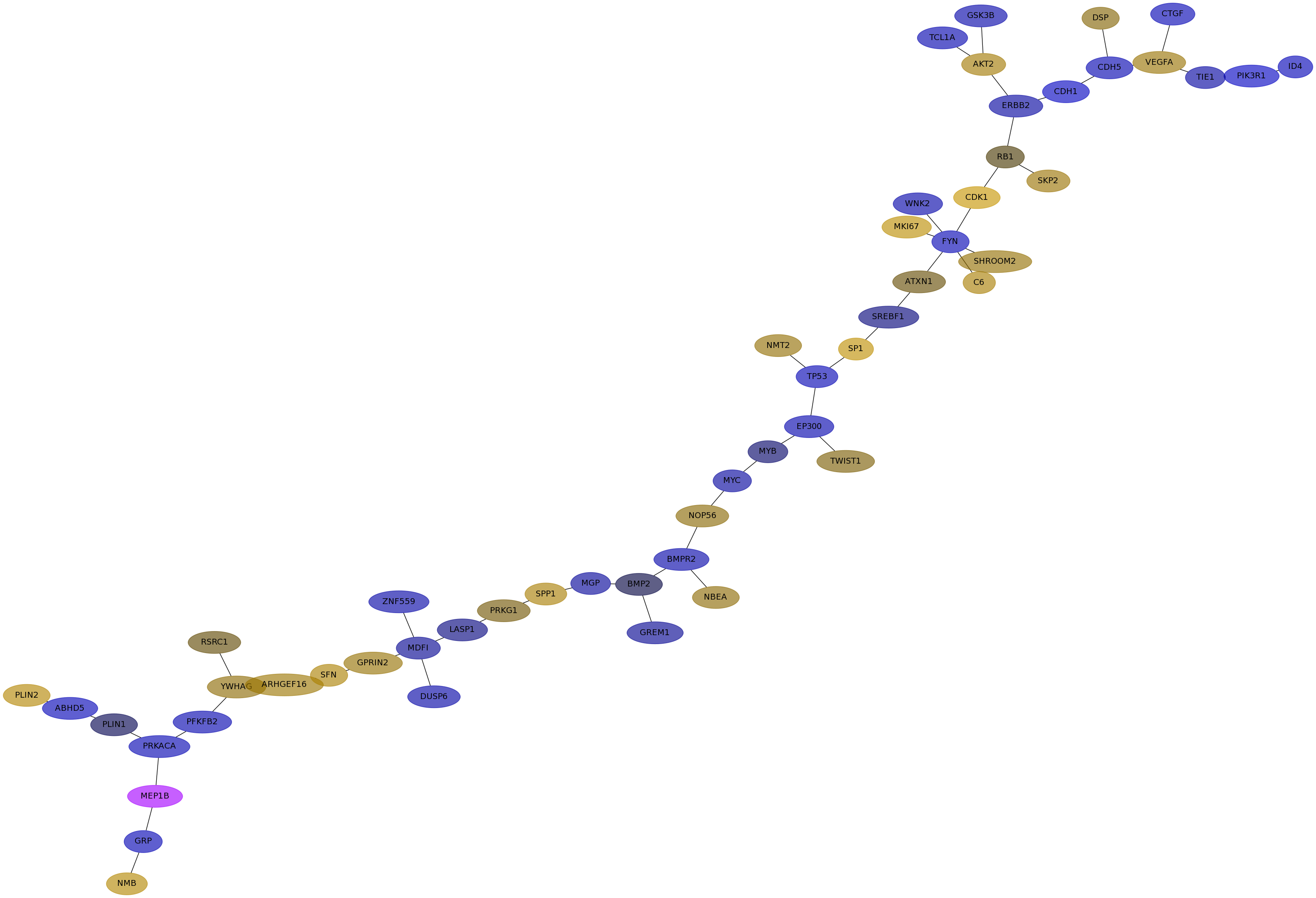
Score for each gene in subnetwork 6666 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| LASP1 |   | 1 | 1195 | 1450 | 1465 | -0.048 | 0.033 | -0.067 | 0.219 | 0.069 |
|---|
| BMPR2 |   | 5 | 360 | 1323 | 1292 | -0.132 | 0.081 | -0.029 | 0.356 | -0.021 |
|---|
| DUSP6 |   | 1 | 1195 | 1450 | 1465 | -0.119 | 0.167 | 0.018 | 0.038 | -0.007 |
|---|
| C6 |   | 4 | 440 | 913 | 904 | 0.128 | 0.055 | 0.169 | 0.161 | 0.054 |
|---|
| MGP |   | 3 | 557 | 1450 | 1430 | -0.088 | 0.007 | 0.150 | -0.084 | -0.030 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| PRKACA |   | 2 | 743 | 478 | 490 | -0.163 | 0.181 | 0.104 | -0.038 | -0.144 |
|---|
| NMB |   | 1 | 1195 | 1450 | 1465 | 0.169 | -0.057 | 0.232 | -0.008 | -0.028 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| ARHGEF16 |   | 10 | 167 | 296 | 283 | 0.096 | 0.036 | -0.047 | 0.200 | 0.038 |
|---|
| NOP56 |   | 3 | 557 | 1450 | 1430 | 0.062 | 0.036 | 0.030 | undef | 0.011 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| PRKG1 |   | 3 | 557 | 179 | 213 | 0.035 | -0.125 | -0.074 | 0.183 | -0.095 |
|---|
| SREBF1 |   | 9 | 196 | 478 | 460 | -0.042 | 0.232 | -0.195 | 0.140 | -0.074 |
|---|
| NBEA |   | 1 | 1195 | 1450 | 1465 | 0.066 | 0.093 | 0.098 | 0.136 | -0.052 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PLIN1 |   | 1 | 1195 | 1450 | 1465 | -0.016 | 0.046 | 0.145 | 0.186 | 0.069 |
|---|
| WNK2 |   | 7 | 256 | 1 | 47 | -0.127 | 0.029 | undef | 0.201 | undef |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| GRP |   | 1 | 1195 | 1450 | 1465 | -0.169 | 0.255 | -0.032 | 0.112 | 0.147 |
|---|
| GREM1 |   | 9 | 196 | 525 | 508 | -0.073 | 0.200 | 0.028 | 0.225 | 0.261 |
|---|
| ZNF559 |   | 3 | 557 | 913 | 908 | -0.122 | 0.089 | 0.145 | -0.005 | -0.014 |
|---|
| PFKFB2 |   | 3 | 557 | 692 | 691 | -0.149 | -0.081 | -0.158 | 0.121 | -0.202 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| MKI67 |   | 5 | 360 | 1 | 56 | 0.205 | -0.123 | 0.176 | -0.086 | 0.018 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| DSP |   | 7 | 256 | 590 | 570 | 0.054 | 0.235 | -0.119 | -0.033 | 0.053 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| NMT2 |   | 7 | 256 | 906 | 876 | 0.077 | -0.296 | 0.134 | -0.124 | 0.217 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| ABHD5 |   | 1 | 1195 | 1450 | 1465 | -0.182 | 0.062 | 0.007 | -0.111 | -0.192 |
|---|
| PLIN2 |   | 1 | 1195 | 1450 | 1465 | 0.166 | -0.014 | 0.136 | 0.080 | 0.149 |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| CDH5 |   | 2 | 743 | 1055 | 1059 | -0.159 | 0.023 | 0.180 | undef | 0.033 |
|---|
| MEP1B |   | 2 | 743 | 913 | 914 | undef | 0.053 | 0.142 | 0.118 | -0.167 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| TIE1 |   | 18 | 86 | 366 | 357 | -0.108 | 0.005 | 0.141 | -0.108 | 0.252 |
|---|
| TCL1A |   | 3 | 557 | 1450 | 1430 | -0.152 | -0.166 | 0.090 | undef | -0.066 |
|---|
| SHROOM2 |   | 3 | 557 | 1396 | 1382 | 0.080 | 0.059 | -0.174 | 0.142 | -0.105 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6666 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.42E-07 | 3.468E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 6.76E-07 | 8.258E-04 |
|---|
| cellular response to starvation | GO:0009267 |  | 1.015E-06 | 8.266E-04 |
|---|
| regulation of bone mineralization | GO:0030500 |  | 1.225E-06 | 7.485E-04 |
|---|
| regulation of biomineral formation | GO:0070167 |  | 1.741E-06 | 8.507E-04 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 1.795E-06 | 7.308E-04 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 1.959E-06 | 6.836E-04 |
|---|
| ER overload response | GO:0006983 |  | 2.347E-06 | 7.168E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.347E-06 | 6.372E-04 |
|---|
| response to starvation | GO:0042594 |  | 2.403E-06 | 5.869E-04 |
|---|
| cellular response to nutrient levels | GO:0031669 |  | 2.403E-06 | 5.336E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 1.7E-07 | 4.091E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 8.09E-07 | 9.732E-04 |
|---|
| regulation of bone mineralization | GO:0030500 |  | 1.466E-06 | 1.176E-03 |
|---|
| regulation of biomineral formation | GO:0070167 |  | 2.082E-06 | 1.253E-03 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 2.232E-06 | 1.074E-03 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 2.435E-06 | 9.766E-04 |
|---|
| ER overload response | GO:0006983 |  | 2.692E-06 | 9.253E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.692E-06 | 8.096E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5E-06 | 1.337E-03 |
|---|
| lung development | GO:0030324 |  | 5E-06 | 1.203E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.381E-06 | 1.177E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 5.262E-07 | 1.21E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.585E-06 | 1.822E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.274E-06 | 1.744E-03 |
|---|
| regulation of bone mineralization | GO:0030500 |  | 3.065E-06 | 1.762E-03 |
|---|
| regulation of biomineral formation | GO:0070167 |  | 4.5E-06 | 2.07E-03 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 6.105E-06 | 2.34E-03 |
|---|
| regulation of gliogenesis | GO:0014013 |  | 6.323E-06 | 2.078E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.323E-06 | 1.818E-03 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 6.702E-06 | 1.713E-03 |
|---|
| lung alveolus development | GO:0048286 |  | 9.451E-06 | 2.174E-03 |
|---|
| lung development | GO:0030324 |  | 1.041E-05 | 2.176E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 1.714E-06 | 3.16E-03 |
|---|
| lung development | GO:0030324 |  | 1.714E-06 | 1.58E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.966E-06 | 1.208E-03 |
|---|
| respiratory tube development | GO:0030323 |  | 2.246E-06 | 1.035E-03 |
|---|
| respiratory system development | GO:0060541 |  | 2.246E-06 | 8.278E-04 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 2.556E-06 | 7.851E-04 |
|---|
| N-terminal protein amino acid modification | GO:0031365 |  | 3.33E-06 | 8.766E-04 |
|---|
| cartilage condensation | GO:0001502 |  | 3.33E-06 | 7.67E-04 |
|---|
| ER overload response | GO:0006983 |  | 3.33E-06 | 6.818E-04 |
|---|
| tube development | GO:0035295 |  | 5.285E-06 | 9.741E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 9.25E-06 | 1.55E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 5.262E-07 | 1.21E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.585E-06 | 1.822E-03 |
|---|
| ER overload response | GO:0006983 |  | 2.274E-06 | 1.744E-03 |
|---|
| regulation of bone mineralization | GO:0030500 |  | 3.065E-06 | 1.762E-03 |
|---|
| regulation of biomineral formation | GO:0070167 |  | 4.5E-06 | 2.07E-03 |
|---|
| regulation of bone remodeling | GO:0046850 |  | 6.105E-06 | 2.34E-03 |
|---|
| regulation of gliogenesis | GO:0014013 |  | 6.323E-06 | 2.078E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 6.323E-06 | 1.818E-03 |
|---|
| regulation of tissue remodeling | GO:0034103 |  | 6.702E-06 | 1.713E-03 |
|---|
| lung alveolus development | GO:0048286 |  | 9.451E-06 | 2.174E-03 |
|---|
| lung development | GO:0030324 |  | 1.041E-05 | 2.176E-03 |
|---|



