Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6635
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7530 | 3.568e-03 | 5.830e-04 | 7.728e-01 | 1.608e-06 |
|---|
| Loi | 0.2305 | 7.998e-02 | 1.128e-02 | 4.275e-01 | 3.858e-04 |
|---|
| Schmidt | 0.6742 | 0.000e+00 | 0.000e+00 | 4.410e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.466e-03 | 0.000e+00 |
|---|
| Wang | 0.2624 | 2.563e-03 | 4.276e-02 | 3.914e-01 | 4.289e-05 |
|---|
Expression data for subnetwork 6635 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
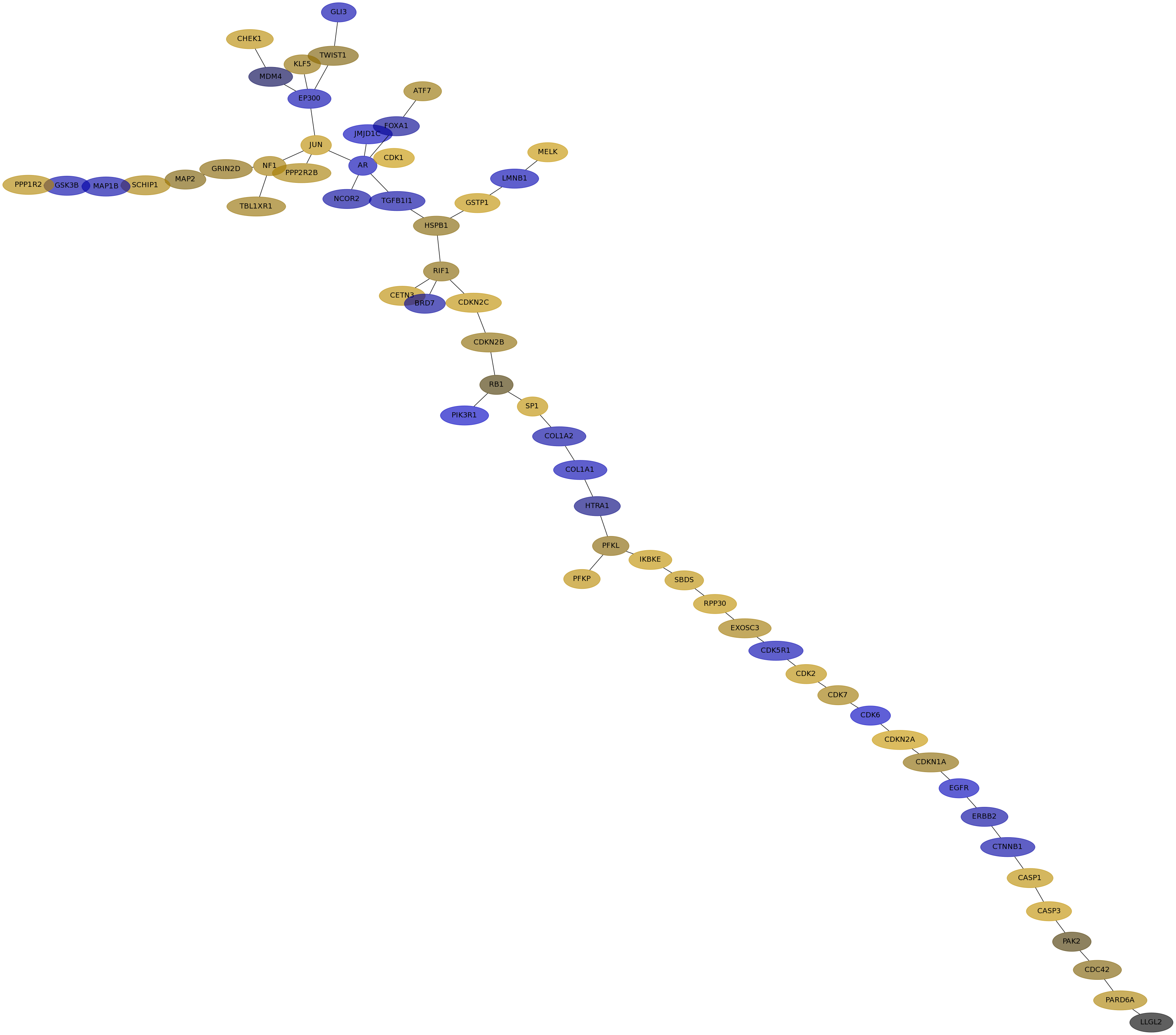
Score for each gene in subnetwork 6635 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| CDKN2B |   | 7 | 256 | 236 | 237 | 0.066 | 0.050 | -0.028 | 0.039 | 0.153 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| CASP3 |   | 11 | 148 | 141 | 143 | 0.234 | -0.032 | -0.142 | -0.097 | -0.084 |
|---|
| PARD6A |   | 2 | 743 | 366 | 395 | 0.140 | -0.084 | 0.008 | 0.161 | 0.084 |
|---|
| PPP1R2 |   | 1 | 1195 | 552 | 606 | 0.163 | 0.000 | 0.035 | -0.038 | 0.037 |
|---|
| EXOSC3 |   | 3 | 557 | 552 | 558 | 0.107 | 0.121 | undef | -0.074 | undef |
|---|
| NF1 |   | 9 | 196 | 552 | 541 | 0.110 | -0.052 | -0.032 | 0.090 | -0.094 |
|---|
| HTRA1 |   | 7 | 256 | 83 | 102 | -0.046 | 0.184 | 0.051 | 0.132 | 0.224 |
|---|
| RIF1 |   | 2 | 743 | 552 | 568 | 0.057 | -0.060 | -0.035 | undef | -0.092 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| MDM4 |   | 9 | 196 | 141 | 147 | -0.016 | 0.004 | 0.164 | 0.107 | 0.105 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CHEK1 |   | 18 | 86 | 141 | 136 | 0.189 | -0.134 | 0.119 | -0.088 | -0.054 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| GSTP1 |   | 14 | 117 | 1 | 34 | 0.222 | -0.166 | 0.201 | 0.126 | -0.136 |
|---|
| SBDS |   | 1 | 1195 | 552 | 606 | 0.204 | -0.063 | undef | 0.075 | undef |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| RPP30 |   | 1 | 1195 | 552 | 606 | 0.217 | -0.091 | 0.171 | -0.053 | -0.065 |
|---|
| CDKN2C |   | 16 | 104 | 236 | 227 | 0.216 | -0.106 | 0.202 | -0.024 | -0.081 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| CTNNB1 |   | 43 | 25 | 83 | 81 | -0.113 | 0.187 | 0.159 | -0.090 | 0.239 |
|---|
| LMNB1 |   | 2 | 743 | 83 | 123 | -0.158 | -0.177 | 0.010 | 0.006 | 0.156 |
|---|
| CDC42 |   | 26 | 52 | 179 | 177 | 0.047 | 0.242 | -0.027 | 0.040 | -0.042 |
|---|
| PFKL |   | 7 | 256 | 83 | 102 | 0.056 | 0.106 | -0.173 | 0.160 | -0.158 |
|---|
| LLGL2 |   | 1 | 1195 | 552 | 606 | -0.000 | 0.209 | -0.030 | 0.178 | -0.062 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| SCHIP1 |   | 10 | 167 | 478 | 459 | 0.130 | 0.136 | 0.105 | -0.022 | 0.198 |
|---|
| BRD7 |   | 23 | 66 | 457 | 437 | -0.090 | 0.019 | -0.188 | undef | -0.086 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| CASP1 |   | 6 | 301 | 552 | 544 | 0.204 | -0.166 | -0.046 | undef | -0.034 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| CETN3 |   | 1 | 1195 | 552 | 606 | 0.199 | 0.089 | 0.108 | -0.026 | 0.155 |
|---|
| TBL1XR1 |   | 3 | 557 | 552 | 558 | 0.084 | -0.064 | undef | undef | undef |
|---|
| COL1A1 |   | 3 | 557 | 552 | 558 | -0.156 | 0.244 | -0.021 | -0.081 | 0.086 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| JMJD1C |   | 7 | 256 | 552 | 542 | -0.195 | 0.241 | 0.074 | undef | 0.129 |
|---|
| CDK7 |   | 7 | 256 | 318 | 328 | 0.105 | 0.184 | 0.017 | 0.078 | -0.097 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| CDK5R1 |   | 17 | 95 | 552 | 521 | -0.150 | 0.038 | -0.096 | 0.121 | 0.091 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| IKBKE |   | 10 | 167 | 552 | 539 | 0.227 | -0.041 | -0.095 | -0.197 | 0.079 |
|---|
| MAP2 |   | 3 | 557 | 552 | 558 | 0.044 | 0.045 | 0.014 | undef | -0.057 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| GRIN2D |   | 13 | 124 | 552 | 522 | 0.058 | 0.155 | 0.000 | undef | undef |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| COL1A2 |   | 7 | 256 | 552 | 542 | -0.110 | 0.207 | 0.046 | 0.235 | 0.097 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| MELK |   | 1 | 1195 | 552 | 606 | 0.253 | -0.187 | 0.031 | 0.000 | 0.060 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
| PFKP |   | 6 | 301 | 83 | 105 | 0.195 | -0.076 | -0.040 | undef | 0.094 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
GO Enrichment output for subnetwork 6635 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.547E-15 | 1.111E-11 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.321E-12 | 2.834E-09 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.634E-09 | 1.331E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.142E-09 | 1.308E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.773E-09 | 1.355E-06 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.677E-08 | 1.09E-05 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 3.914E-08 | 1.366E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 3.914E-08 | 1.195E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.187E-08 | 1.408E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.739E-08 | 1.402E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 8.474E-08 | 1.882E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.004E-14 | 2.415E-11 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.574E-12 | 4.3E-09 |
|---|
| interphase | GO:0051325 |  | 5.06E-12 | 4.058E-09 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 2.544E-09 | 1.53E-06 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 3.333E-09 | 1.604E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 4.314E-09 | 1.73E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 2.652E-08 | 9.115E-06 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 3.493E-08 | 1.051E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 5.282E-08 | 1.412E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 6.967E-08 | 1.676E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 7.732E-08 | 1.691E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.105E-13 | 9.441E-10 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.563E-11 | 1.1E-07 |
|---|
| interphase | GO:0051325 |  | 1.207E-10 | 9.253E-08 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.776E-09 | 2.171E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 6.56E-09 | 3.017E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.241E-08 | 1.626E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.759E-08 | 1.564E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.385E-08 | 1.548E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 7.7E-08 | 1.968E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 8.443E-08 | 1.942E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 8.443E-08 | 1.765E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.081E-10 | 7.521E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 9.276E-10 | 8.547E-07 |
|---|
| androgen receptor signaling pathway | GO:0030521 |  | 2.285E-07 | 1.404E-04 |
|---|
| gland development | GO:0048732 |  | 3.544E-07 | 1.633E-04 |
|---|
| G1 phase | GO:0051318 |  | 5.192E-07 | 1.914E-04 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 6.247E-07 | 1.919E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 7.467E-07 | 1.966E-04 |
|---|
| prostate gland development | GO:0030850 |  | 1.949E-06 | 4.489E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.214E-06 | 4.534E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.538E-06 | 4.678E-04 |
|---|
| urogenital system development | GO:0001655 |  | 2.538E-06 | 4.253E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.105E-13 | 9.441E-10 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 9.563E-11 | 1.1E-07 |
|---|
| interphase | GO:0051325 |  | 1.207E-10 | 9.253E-08 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 3.776E-09 | 2.171E-06 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 6.56E-09 | 3.017E-06 |
|---|
| regulation of lipid kinase activity | GO:0043550 |  | 4.241E-08 | 1.626E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 4.759E-08 | 1.564E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.385E-08 | 1.548E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 7.7E-08 | 1.968E-05 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 8.443E-08 | 1.942E-05 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 8.443E-08 | 1.765E-05 |
|---|



