Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6586
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7542 | 3.493e-03 | 5.670e-04 | 7.698e-01 | 1.525e-06 |
|---|
| Loi | 0.2305 | 7.993e-02 | 1.127e-02 | 4.274e-01 | 3.851e-04 |
|---|
| Schmidt | 0.6735 | 0.000e+00 | 0.000e+00 | 4.479e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.471e-03 | 0.000e+00 |
|---|
| Wang | 0.2620 | 2.627e-03 | 4.335e-02 | 3.942e-01 | 4.489e-05 |
|---|
Expression data for subnetwork 6586 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
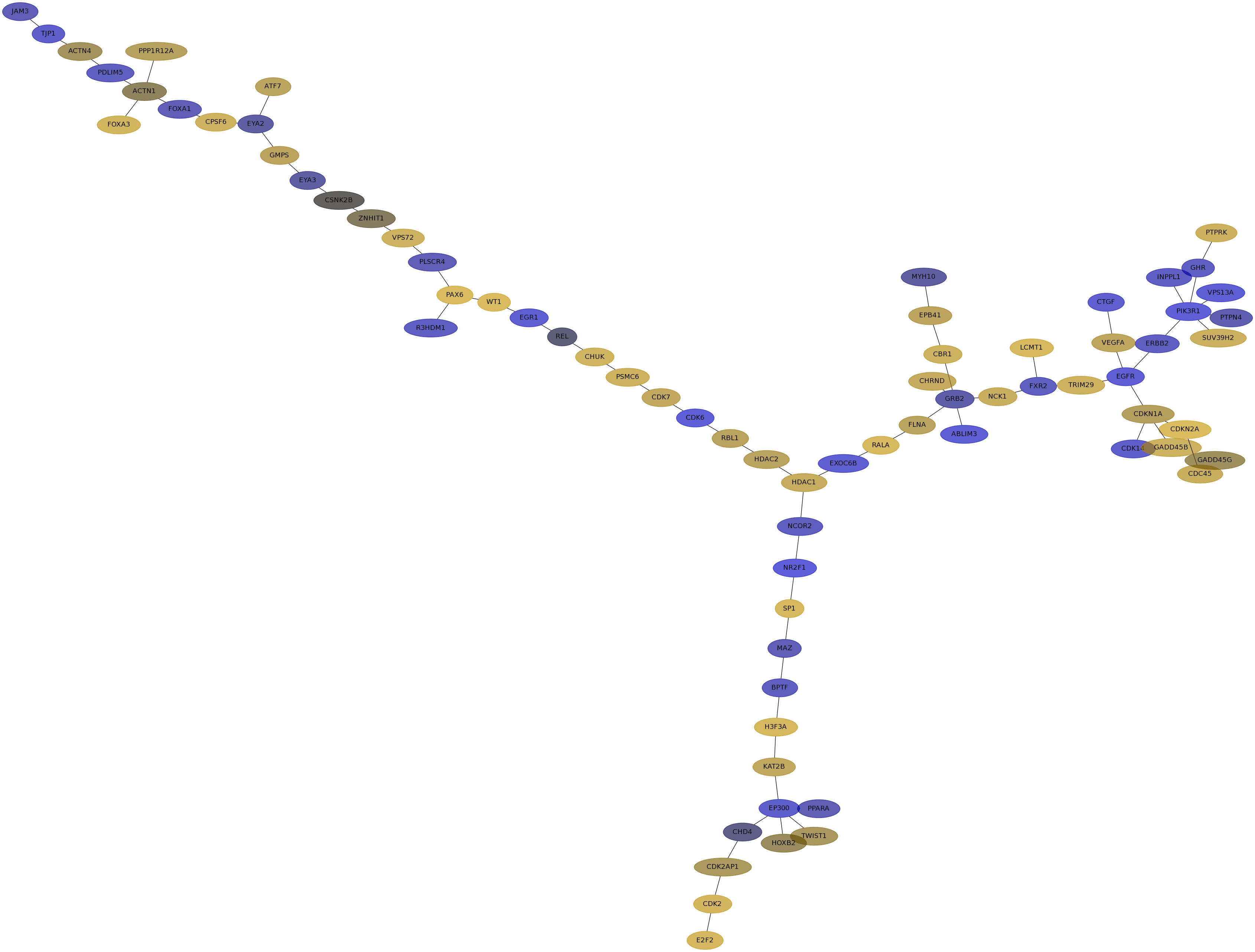
Score for each gene in subnetwork 6586 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| BPTF |   | 4 | 440 | 318 | 337 | -0.098 | 0.159 | 0.132 | undef | 0.030 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CSNK2B |   | 3 | 557 | 318 | 341 | 0.001 | -0.039 | -0.004 | 0.115 | 0.033 |
|---|
| CHUK |   | 6 | 301 | 318 | 330 | 0.173 | 0.080 | 0.152 | undef | 0.215 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| VPS13A |   | 10 | 167 | 236 | 234 | -0.203 | 0.105 | -0.046 | 0.120 | 0.062 |
|---|
| MYH10 |   | 5 | 360 | 236 | 242 | -0.029 | 0.139 | 0.132 | undef | 0.223 |
|---|
| CDK14 |   | 7 | 256 | 179 | 201 | -0.151 | 0.011 | -0.071 | 0.212 | 0.107 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| CHD4 |   | 2 | 743 | 318 | 346 | -0.012 | 0.079 | 0.092 | 0.062 | -0.070 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| CPSF6 |   | 6 | 301 | 318 | 330 | 0.164 | 0.052 | 0.091 | 0.129 | -0.088 |
|---|
| CDK6 |   | 16 | 104 | 318 | 314 | -0.229 | 0.107 | -0.028 | -0.118 | 0.104 |
|---|
| PTPRK |   | 14 | 117 | 83 | 96 | 0.156 | 0.141 | -0.094 | 0.048 | -0.024 |
|---|
| NCK1 |   | 3 | 557 | 318 | 341 | 0.135 | -0.100 | -0.059 | -0.108 | 0.151 |
|---|
| EXOC6B |   | 6 | 301 | 318 | 330 | -0.183 | 0.205 | -0.104 | 0.030 | 0.037 |
|---|
| EYA3 |   | 9 | 196 | 318 | 324 | -0.034 | 0.053 | -0.020 | 0.188 | -0.142 |
|---|
| R3HDM1 |   | 4 | 440 | 318 | 337 | -0.108 | -0.192 | -0.049 | 0.082 | 0.223 |
|---|
| ACTN1 |   | 18 | 86 | 318 | 313 | 0.016 | 0.347 | 0.068 | 0.100 | 0.314 |
|---|
| FLNA |   | 50 | 20 | 1 | 14 | 0.081 | 0.107 | 0.148 | 0.268 | 0.126 |
|---|
| CDK7 |   | 7 | 256 | 318 | 328 | 0.105 | 0.184 | 0.017 | 0.078 | -0.097 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| JAM3 |   | 1 | 1195 | 318 | 382 | -0.068 | 0.218 | 0.153 | undef | 0.155 |
|---|
| EPB41 |   | 2 | 743 | 318 | 346 | 0.082 | -0.247 | 0.000 | -0.179 | -0.042 |
|---|
| CDC45 |   | 5 | 360 | 318 | 334 | 0.134 | -0.171 | 0.185 | -0.103 | -0.085 |
|---|
| RALA |   | 10 | 167 | 318 | 323 | 0.224 | 0.207 | -0.047 | 0.169 | 0.146 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| PSMC6 |   | 5 | 360 | 318 | 334 | 0.163 | 0.061 | -0.010 | 0.069 | 0.044 |
|---|
| GADD45B |   | 8 | 222 | 318 | 325 | 0.158 | 0.122 | 0.067 | 0.060 | 0.212 |
|---|
| HDAC1 |   | 4 | 440 | 318 | 337 | 0.122 | -0.020 | 0.050 | -0.035 | -0.172 |
|---|
| ACTN4 |   | 14 | 117 | 318 | 315 | 0.035 | 0.180 | undef | -0.048 | undef |
|---|
| H3F3A |   | 2 | 743 | 318 | 346 | 0.221 | 0.232 | 0.151 | 0.237 | -0.142 |
|---|
| E2F2 |   | 13 | 124 | 1 | 35 | 0.212 | -0.047 | 0.162 | -0.131 | -0.092 |
|---|
| EYA2 |   | 13 | 124 | 318 | 316 | -0.034 | 0.113 | -0.142 | undef | -0.019 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
| TRIM29 |   | 8 | 222 | 296 | 287 | 0.161 | 0.240 | -0.150 | -0.077 | 0.024 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| INPPL1 |   | 6 | 301 | 141 | 152 | -0.122 | 0.072 | -0.087 | 0.174 | 0.156 |
|---|
| PLSCR4 |   | 13 | 124 | 318 | 316 | -0.069 | 0.140 | 0.231 | 0.212 | 0.212 |
|---|
| ZNHIT1 |   | 2 | 743 | 318 | 346 | 0.010 | 0.095 | -0.047 | 0.030 | 0.072 |
|---|
| EGR1 |   | 39 | 29 | 1 | 18 | -0.184 | 0.259 | 0.130 | 0.018 | 0.122 |
|---|
| KAT2B |   | 13 | 124 | 318 | 316 | 0.102 | -0.157 | -0.091 | 0.136 | -0.087 |
|---|
| PPP1R12A |   | 2 | 743 | 318 | 346 | 0.069 | 0.124 | 0.114 | 0.100 | 0.018 |
|---|
| CHRND |   | 3 | 557 | 318 | 341 | 0.114 | -0.031 | -0.047 | 0.154 | -0.031 |
|---|
| REL |   | 2 | 743 | 318 | 346 | -0.006 | -0.018 | -0.101 | -0.093 | -0.183 |
|---|
| ATF7 |   | 28 | 47 | 179 | 175 | 0.085 | 0.115 | -0.088 | 0.172 | -0.092 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| FOXA3 |   | 12 | 140 | 318 | 322 | 0.192 | 0.289 | undef | -0.171 | undef |
|---|
| TJP1 |   | 8 | 222 | 318 | 325 | -0.139 | 0.102 | 0.099 | 0.319 | 0.049 |
|---|
| HOXB2 |   | 33 | 37 | 318 | 289 | 0.024 | -0.143 | 0.076 | 0.247 | -0.184 |
|---|
| SUV39H2 |   | 8 | 222 | 318 | 325 | 0.150 | -0.097 | -0.057 | undef | 0.028 |
|---|
| VPS72 |   | 6 | 301 | 318 | 330 | 0.164 | -0.182 | 0.095 | 0.151 | 0.080 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| GMPS |   | 7 | 256 | 318 | 328 | 0.083 | -0.022 | 0.044 | -0.019 | 0.151 |
|---|
| WT1 |   | 33 | 37 | 1 | 21 | 0.254 | -0.039 | 0.036 | undef | 0.172 |
|---|
| ABLIM3 |   | 1 | 1195 | 318 | 382 | -0.196 | 0.233 | -0.010 | -0.033 | 0.150 |
|---|
| MAZ |   | 2 | 743 | 318 | 346 | -0.068 | -0.208 | -0.157 | undef | -0.175 |
|---|
| CDK2AP1 |   | 1 | 1195 | 318 | 382 | 0.048 | 0.067 | 0.138 | 0.128 | -0.151 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| PPARA |   | 1 | 1195 | 318 | 382 | -0.067 | -0.101 | 0.005 | -0.031 | -0.111 |
|---|
| FXR2 |   | 1 | 1195 | 318 | 382 | -0.092 | -0.113 | -0.082 | -0.058 | 0.107 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| NR2F1 |   | 25 | 59 | 83 | 92 | -0.252 | 0.113 | 0.133 | -0.116 | 0.207 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| LCMT1 |   | 1 | 1195 | 318 | 382 | 0.225 | -0.209 | -0.010 | undef | -0.242 |
|---|
| CBR1 |   | 1 | 1195 | 318 | 382 | 0.160 | -0.252 | 0.052 | -0.162 | -0.050 |
|---|
| PTPN4 |   | 2 | 743 | 318 | 346 | -0.055 | -0.110 | -0.066 | 0.013 | 0.083 |
|---|
| PDLIM5 |   | 4 | 440 | 318 | 337 | -0.091 | 0.242 | -0.085 | -0.001 | 0.117 |
|---|
| CDK2 |   | 31 | 43 | 179 | 174 | 0.196 | -0.096 | 0.109 | 0.001 | 0.362 |
|---|
| CDKN2A |   | 60 | 17 | 1 | 11 | 0.263 | -0.113 | 0.111 | undef | -0.105 |
|---|
| FOXA1 |   | 27 | 50 | 179 | 176 | -0.072 | 0.071 | -0.015 | 0.214 | 0.005 |
|---|
GO Enrichment output for subnetwork 6586 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| histone modification | GO:0016570 |  | 4.042E-07 | 9.875E-04 |
|---|
| covalent chromatin modification | GO:0016569 |  | 5.082E-07 | 6.207E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.177E-06 | 9.588E-04 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 3.163E-06 | 1.932E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 5.514E-06 | 2.694E-03 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 7.217E-06 | 2.939E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 8.787E-06 | 3.067E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 8.787E-06 | 2.683E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 1.067E-05 | 2.897E-03 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 1.067E-05 | 2.607E-03 |
|---|
| vesicle docking during exocytosis | GO:0006904 |  | 1.171E-05 | 2.6E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| histone modification | GO:0016570 |  | 4.39E-07 | 1.056E-03 |
|---|
| covalent chromatin modification | GO:0016569 |  | 5.571E-07 | 6.702E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.372E-06 | 1.1E-03 |
|---|
| interphase | GO:0051325 |  | 1.692E-06 | 1.018E-03 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 3.743E-06 | 1.801E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 6.522E-06 | 2.615E-03 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 8.509E-06 | 2.925E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.039E-05 | 3.125E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.039E-05 | 2.778E-03 |
|---|
| chromatin remodeling | GO:0006338 |  | 1.151E-05 | 2.77E-03 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 1.151E-05 | 2.518E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| histone modification | GO:0016570 |  | 6.572E-07 | 1.511E-03 |
|---|
| covalent chromatin modification | GO:0016569 |  | 7.492E-07 | 8.615E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.953E-06 | 1.497E-03 |
|---|
| interphase | GO:0051325 |  | 2.282E-06 | 1.312E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 6.06E-06 | 2.787E-03 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 6.06E-06 | 2.323E-03 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 1.18E-05 | 3.876E-03 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 1.639E-05 | 4.713E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.68E-05 | 4.293E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.68E-05 | 3.864E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.68E-05 | 3.512E-03 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| actin cytoskeleton reorganization | GO:0031532 |  | 3.798E-06 | 7E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.327E-05 | 0.03065856 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 3.672E-05 | 0.02256075 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 4.186E-05 | 0.01928543 |
|---|
| histone deacetylation | GO:0016575 |  | 4.44E-05 | 0.01636636 |
|---|
| histone modification | GO:0016570 |  | 4.81E-05 | 0.01477477 |
|---|
| regulation of epithelial cell proliferation | GO:0050678 |  | 5.549E-05 | 0.01461023 |
|---|
| covalent chromatin modification | GO:0016569 |  | 5.549E-05 | 0.01278395 |
|---|
| G1 phase of mitotic cell cycle | GO:0000080 |  | 6.073E-05 | 0.01243675 |
|---|
| signal complex assembly | GO:0007172 |  | 6.073E-05 | 0.01119307 |
|---|
| activation of MAPKK activity | GO:0000186 |  | 6.073E-05 | 0.01017552 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| histone modification | GO:0016570 |  | 6.572E-07 | 1.511E-03 |
|---|
| covalent chromatin modification | GO:0016569 |  | 7.492E-07 | 8.615E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.953E-06 | 1.497E-03 |
|---|
| interphase | GO:0051325 |  | 2.282E-06 | 1.312E-03 |
|---|
| N-terminal protein amino acid acetylation | GO:0006474 |  | 6.06E-06 | 2.787E-03 |
|---|
| activation of MAPKKK activity | GO:0000185 |  | 6.06E-06 | 2.323E-03 |
|---|
| regulation of mast cell proliferation | GO:0070666 |  | 1.18E-05 | 3.876E-03 |
|---|
| regulation of leukocyte proliferation | GO:0070663 |  | 1.639E-05 | 4.713E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 1.68E-05 | 4.293E-03 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 1.68E-05 | 3.864E-03 |
|---|
| regulation of T cell differentiation in the thymus | GO:0033081 |  | 1.68E-05 | 3.512E-03 |
|---|



