Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6573
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7543 | 3.491e-03 | 5.670e-04 | 7.697e-01 | 1.523e-06 |
|---|
| Loi | 0.2305 | 7.992e-02 | 1.127e-02 | 4.274e-01 | 3.848e-04 |
|---|
| Schmidt | 0.6733 | 0.000e+00 | 0.000e+00 | 4.502e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.470e-03 | 0.000e+00 |
|---|
| Wang | 0.2619 | 2.639e-03 | 4.346e-02 | 3.947e-01 | 4.527e-05 |
|---|
Expression data for subnetwork 6573 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
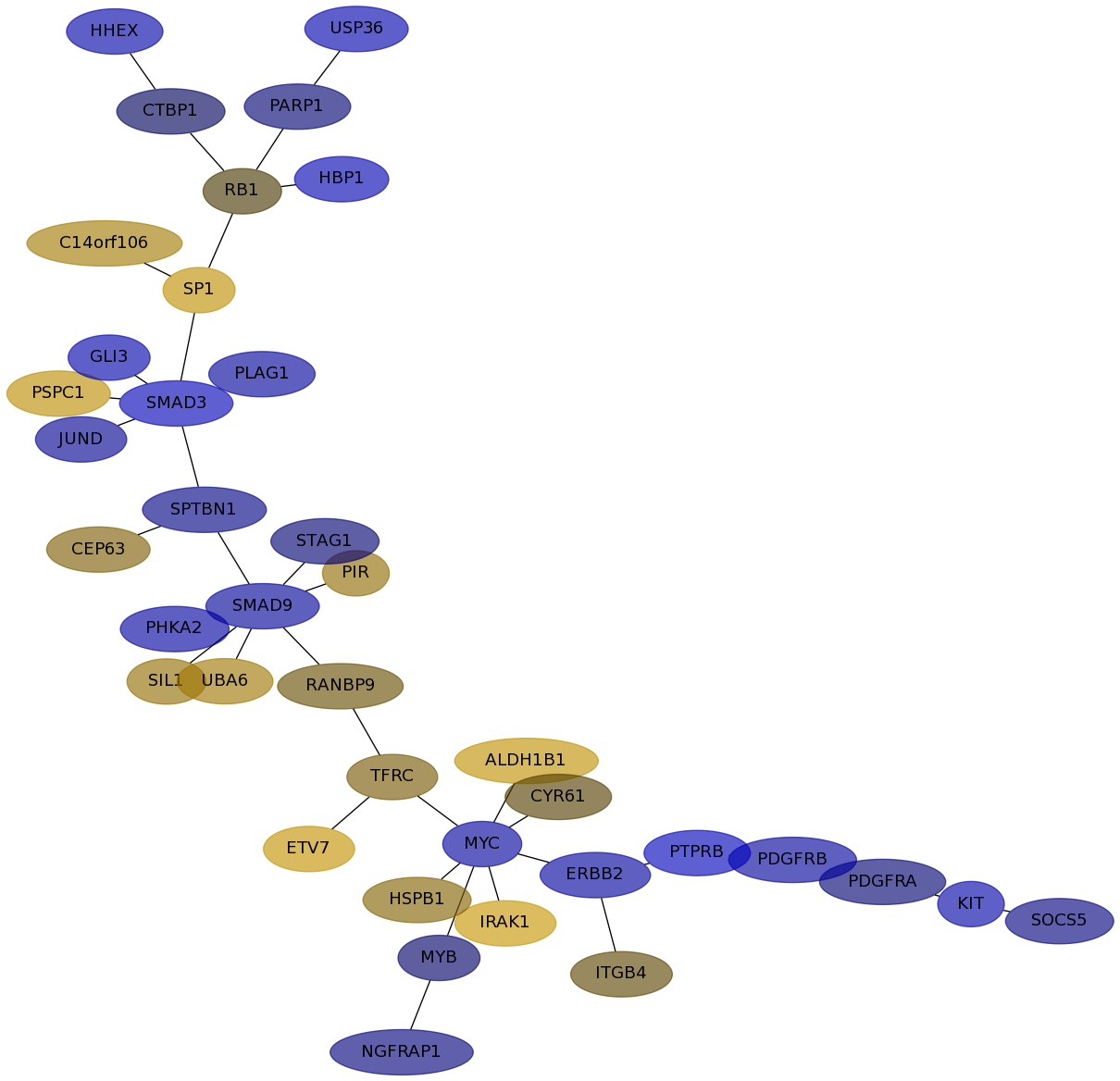
Score for each gene in subnetwork 6573 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| PLAG1 |   | 1 | 1195 | 991 | 1022 | -0.092 | -0.040 | 0.304 | -0.168 | 0.021 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| PDGFRA |   | 10 | 167 | 141 | 146 | -0.035 | 0.008 | 0.118 | 0.107 | 0.119 |
|---|
| C14orf106 |   | 1 | 1195 | 991 | 1022 | 0.112 | -0.203 | 0.208 | -0.006 | -0.229 |
|---|
| TFRC |   | 5 | 360 | 957 | 938 | 0.038 | -0.066 | 0.076 | -0.108 | -0.014 |
|---|
| JUND |   | 2 | 743 | 991 | 999 | -0.077 | 0.091 | -0.075 | 0.007 | -0.005 |
|---|
| RB1 |   | 31 | 43 | 1 | 22 | 0.013 | -0.051 | 0.057 | 0.100 | -0.082 |
|---|
| HBP1 |   | 1 | 1195 | 991 | 1022 | -0.160 | 0.101 | -0.089 | 0.199 | 0.077 |
|---|
| PIR |   | 1 | 1195 | 991 | 1022 | 0.073 | -0.126 | -0.107 | 0.131 | -0.050 |
|---|
| GLI3 |   | 21 | 73 | 478 | 456 | -0.138 | 0.080 | -0.007 | 0.282 | -0.018 |
|---|
| PTPRB |   | 5 | 360 | 991 | 977 | -0.199 | 0.006 | 0.149 | 0.094 | 0.131 |
|---|
| IRAK1 |   | 2 | 743 | 366 | 395 | 0.266 | -0.023 | 0.092 | -0.023 | -0.102 |
|---|
| CYR61 |   | 4 | 440 | 991 | 978 | 0.018 | 0.206 | 0.203 | 0.092 | 0.234 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| SPTBN1 |   | 1 | 1195 | 991 | 1022 | -0.057 | -0.123 | 0.290 | 0.091 | 0.019 |
|---|
| CEP63 |   | 1 | 1195 | 991 | 1022 | 0.046 | 0.181 | 0.108 | undef | 0.231 |
|---|
| PHKA2 |   | 3 | 557 | 991 | 982 | -0.107 | 0.059 | -0.153 | 0.250 | 0.041 |
|---|
| ETV7 |   | 4 | 440 | 957 | 942 | 0.245 | -0.201 | -0.043 | -0.062 | -0.174 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| PDGFRB |   | 12 | 140 | 141 | 142 | -0.094 | 0.224 | 0.127 | 0.153 | 0.189 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| MYB |   | 28 | 47 | 366 | 344 | -0.028 | 0.075 | 0.002 | 0.065 | -0.012 |
|---|
| SOCS5 |   | 1 | 1195 | 991 | 1022 | -0.046 | 0.082 | 0.227 | undef | 0.129 |
|---|
| SMAD9 |   | 9 | 196 | 296 | 285 | -0.090 | -0.153 | 0.055 | 0.081 | -0.059 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| STAG1 |   | 3 | 557 | 991 | 982 | -0.035 | 0.153 | 0.310 | 0.048 | 0.208 |
|---|
| PSPC1 |   | 6 | 301 | 991 | 964 | 0.203 | -0.289 | 0.298 | 0.148 | -0.107 |
|---|
| ALDH1B1 |   | 1 | 1195 | 991 | 1022 | 0.224 | -0.158 | 0.108 | 0.061 | 0.043 |
|---|
| UBA6 |   | 1 | 1195 | 991 | 1022 | 0.105 | 0.028 | -0.040 | 0.244 | -0.040 |
|---|
| RANBP9 |   | 2 | 743 | 991 | 999 | 0.028 | 0.186 | 0.063 | 0.205 | 0.116 |
|---|
| USP36 |   | 3 | 557 | 882 | 871 | -0.144 | -0.044 | 0.203 | undef | 0.014 |
|---|
| SMAD3 |   | 3 | 557 | 842 | 834 | -0.176 | 0.063 | 0.239 | -0.068 | -0.012 |
|---|
| HHEX |   | 4 | 440 | 727 | 723 | -0.130 | -0.178 | 0.177 | -0.180 | 0.005 |
|---|
| NGFRAP1 |   | 3 | 557 | 991 | 982 | -0.044 | 0.005 | 0.252 | -0.001 | 0.109 |
|---|
| SIL1 |   | 1 | 1195 | 991 | 1022 | 0.074 | 0.109 | -0.083 | undef | -0.037 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| HSPB1 |   | 22 | 69 | 1 | 25 | 0.053 | 0.169 | 0.017 | 0.179 | 0.161 |
|---|
| CTBP1 |   | 3 | 557 | 991 | 982 | -0.022 | -0.021 | -0.298 | 0.196 | -0.143 |
|---|
GO Enrichment output for subnetwork 6573 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of alpha-beta T cell activation | GO:0046636 |  | 2.716E-07 | 6.635E-04 |
|---|
| negative regulation of T cell activation | GO:0050868 |  | 3.761E-07 | 4.594E-04 |
|---|
| positive regulation of T-helper 1 cell differentiation | GO:0045627 |  | 5.42E-07 | 4.414E-04 |
|---|
| negative regulation of T cell differentiation | GO:0045581 |  | 5.42E-07 | 3.31E-04 |
|---|
| regulation of T-helper 1 cell differentiation | GO:0045625 |  | 9.464E-07 | 4.624E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 1.226E-06 | 4.99E-04 |
|---|
| negative regulation of lymphocyte activation | GO:0051250 |  | 1.34E-06 | 4.678E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.559E-06 | 4.762E-04 |
|---|
| regulation of T-helper 1 type immune response | GO:0002825 |  | 2.261E-06 | 6.138E-04 |
|---|
| negative regulation of lymphocyte differentiation | GO:0045620 |  | 2.261E-06 | 5.524E-04 |
|---|
| negative regulation of leukocyte activation | GO:0002695 |  | 2.381E-06 | 5.287E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of alpha-beta T cell activation | GO:0046636 |  | 3.643E-07 | 8.765E-04 |
|---|
| negative regulation of T cell activation | GO:0050868 |  | 5.539E-07 | 6.663E-04 |
|---|
| negative regulation of T cell differentiation | GO:0045581 |  | 7.268E-07 | 5.829E-04 |
|---|
| positive regulation of T-helper 1 cell differentiation | GO:0045627 |  | 7.268E-07 | 4.372E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 1.664E-06 | 8.009E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.811E-06 | 7.262E-04 |
|---|
| negative regulation of lymphocyte activation | GO:0051250 |  | 1.97E-06 | 6.772E-04 |
|---|
| regulation of T-helper 1 type immune response | GO:0002825 |  | 2.025E-06 | 6.091E-04 |
|---|
| negative regulation of lymphocyte differentiation | GO:0045620 |  | 3.03E-06 | 8.101E-04 |
|---|
| negative regulation of leukocyte activation | GO:0002695 |  | 3.053E-06 | 7.346E-04 |
|---|
| regulation of T-helper 2 cell differentiation | GO:0045628 |  | 5.923E-06 | 1.296E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of alpha-beta T cell activation | GO:0046636 |  | 4.655E-07 | 1.071E-03 |
|---|
| negative regulation of T cell activation | GO:0050868 |  | 4.809E-07 | 5.53E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 6.12E-07 | 4.692E-04 |
|---|
| negative regulation of T cell differentiation | GO:0045581 |  | 9.286E-07 | 5.339E-04 |
|---|
| positive regulation of T-helper 1 cell differentiation | GO:0045627 |  | 9.286E-07 | 4.272E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 1.545E-06 | 5.921E-04 |
|---|
| negative regulation of lymphocyte activation | GO:0051250 |  | 1.95E-06 | 6.407E-04 |
|---|
| regulation of T-helper 1 type immune response | GO:0002825 |  | 2.586E-06 | 7.436E-04 |
|---|
| negative regulation of leukocyte activation | GO:0002695 |  | 2.691E-06 | 6.876E-04 |
|---|
| negative regulation of lymphocyte differentiation | GO:0045620 |  | 3.87E-06 | 8.9E-04 |
|---|
| negative regulation of cell activation | GO:0050866 |  | 4.167E-06 | 8.713E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.509E-08 | 1.568E-04 |
|---|
| platelet-derived growth factor receptor signaling pathway | GO:0048008 |  | 5.562E-06 | 5.125E-03 |
|---|
| positive regulation of protein import into nucleus | GO:0042307 |  | 5.562E-06 | 3.417E-03 |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.086E-05 | 5.005E-03 |
|---|
| tube development | GO:0035295 |  | 1.9E-05 | 7.005E-03 |
|---|
| protein import into nucleus. translocation | GO:0000060 |  | 2.962E-05 | 9.098E-03 |
|---|
| odontogenesis of dentine-containing tooth | GO:0042475 |  | 2.962E-05 | 7.798E-03 |
|---|
| transmembrane receptor protein serine/threonine kinase signaling pathway | GO:0007178 |  | 4.007E-05 | 9.231E-03 |
|---|
| gland development | GO:0048732 |  | 4.732E-05 | 9.691E-03 |
|---|
| negative regulation of transcription from RNA polymerase II promoter | GO:0000122 |  | 5.282E-05 | 9.735E-03 |
|---|
| peptidyl-tyrosine phosphorylation | GO:0018108 |  | 6.237E-05 | 0.01044913 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of alpha-beta T cell activation | GO:0046636 |  | 4.655E-07 | 1.071E-03 |
|---|
| negative regulation of T cell activation | GO:0050868 |  | 4.809E-07 | 5.53E-04 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 6.12E-07 | 4.692E-04 |
|---|
| negative regulation of T cell differentiation | GO:0045581 |  | 9.286E-07 | 5.339E-04 |
|---|
| positive regulation of T-helper 1 cell differentiation | GO:0045627 |  | 9.286E-07 | 4.272E-04 |
|---|
| cytokine-mediated signaling pathway | GO:0019221 |  | 1.545E-06 | 5.921E-04 |
|---|
| negative regulation of lymphocyte activation | GO:0051250 |  | 1.95E-06 | 6.407E-04 |
|---|
| regulation of T-helper 1 type immune response | GO:0002825 |  | 2.586E-06 | 7.436E-04 |
|---|
| negative regulation of leukocyte activation | GO:0002695 |  | 2.691E-06 | 6.876E-04 |
|---|
| negative regulation of lymphocyte differentiation | GO:0045620 |  | 3.87E-06 | 8.9E-04 |
|---|
| negative regulation of cell activation | GO:0050866 |  | 4.167E-06 | 8.713E-04 |
|---|



