Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6561
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7541 | 3.502e-03 | 5.690e-04 | 7.701e-01 | 1.535e-06 |
|---|
| Loi | 0.2308 | 7.956e-02 | 1.117e-02 | 4.265e-01 | 3.791e-04 |
|---|
| Schmidt | 0.6729 | 0.000e+00 | 0.000e+00 | 4.542e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7908 | 0.000e+00 | 0.000e+00 | 3.459e-03 | 0.000e+00 |
|---|
| Wang | 0.2618 | 2.649e-03 | 4.355e-02 | 3.951e-01 | 4.558e-05 |
|---|
Expression data for subnetwork 6561 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
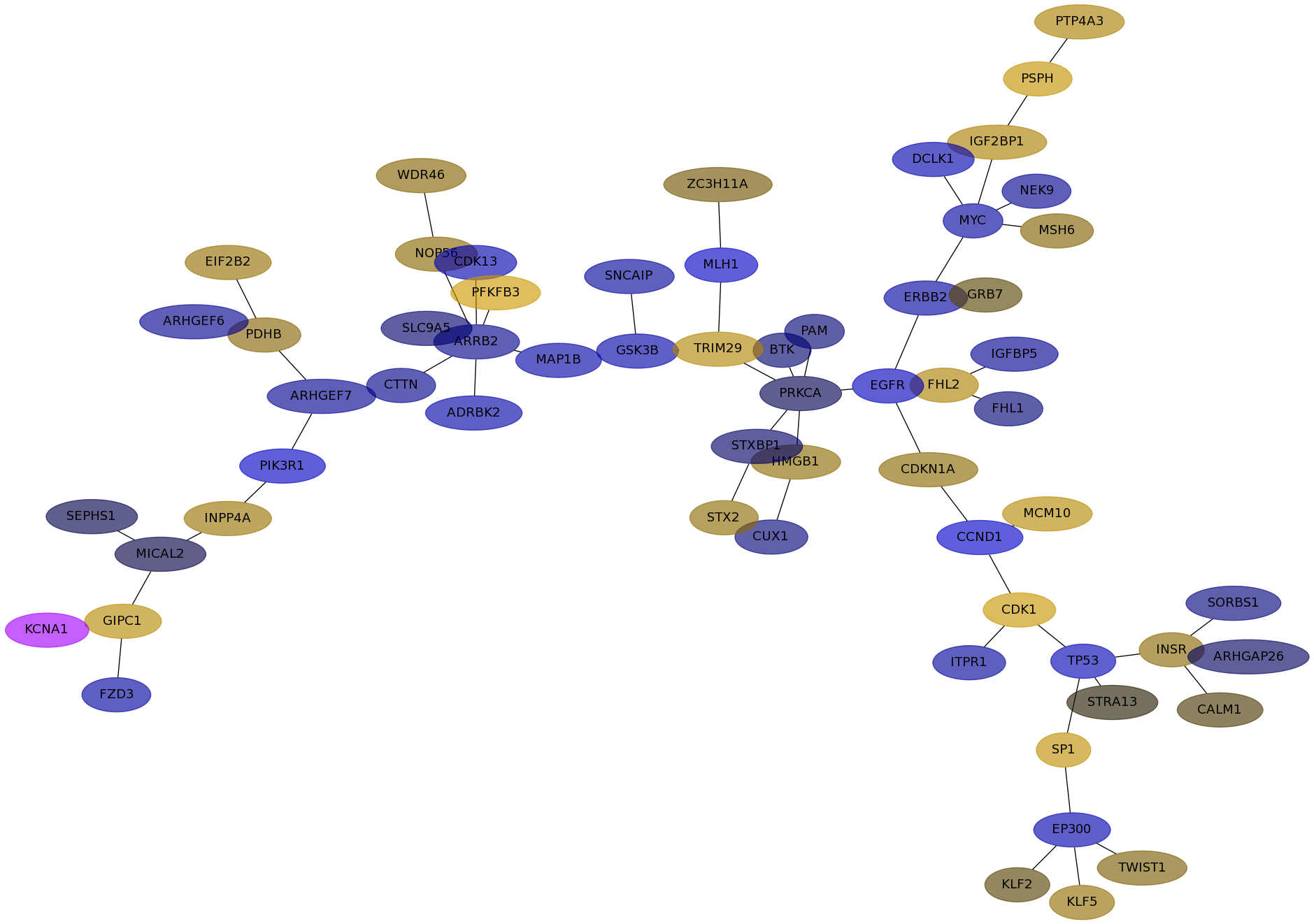
Score for each gene in subnetwork 6561 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| STX2 |   | 1 | 1195 | 1659 | 1678 | 0.070 | -0.115 | 0.143 | undef | 0.274 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| KLF2 |   | 1 | 1195 | 1659 | 1678 | 0.019 | 0.067 | 0.097 | 0.081 | 0.120 |
|---|
| HMGB1 |   | 1 | 1195 | 1659 | 1678 | 0.069 | -0.220 | 0.251 | -0.008 | -0.176 |
|---|
| MCM10 |   | 5 | 360 | 179 | 208 | 0.178 | -0.245 | 0.015 | undef | -0.063 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| STRA13 |   | 1 | 1195 | 1659 | 1678 | 0.005 | -0.181 | 0.135 | undef | 0.017 |
|---|
| PTP4A3 |   | 9 | 196 | 412 | 411 | 0.137 | -0.196 | 0.085 | undef | -0.004 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDK13 |   | 1 | 1195 | 1659 | 1678 | -0.149 | 0.289 | -0.118 | 0.096 | 0.210 |
|---|
| GIPC1 |   | 4 | 440 | 1396 | 1379 | 0.185 | 0.124 | -0.045 | -0.095 | 0.006 |
|---|
| MLH1 |   | 3 | 557 | 1364 | 1342 | -0.237 | 0.122 | 0.032 | 0.020 | -0.035 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| NOP56 |   | 3 | 557 | 1450 | 1430 | 0.062 | 0.036 | 0.030 | undef | 0.011 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| INPP4A |   | 1 | 1195 | 1659 | 1678 | 0.091 | -0.091 | 0.027 | 0.034 | -0.027 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| KCNA1 |   | 2 | 743 | 1659 | 1643 | undef | 0.024 | 0.042 | -0.134 | undef |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| IGF2BP1 |   | 15 | 111 | 366 | 359 | 0.135 | -0.036 | undef | 0.190 | undef |
|---|
| CTTN |   | 2 | 743 | 1247 | 1238 | -0.063 | 0.232 | -0.064 | undef | 0.151 |
|---|
| STXBP1 |   | 5 | 360 | 1629 | 1587 | -0.027 | 0.078 | 0.180 | 0.143 | 0.043 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| SP1 |   | 72 | 10 | 83 | 77 | 0.222 | -0.254 | 0.239 | 0.020 | 0.082 |
|---|
| SEPHS1 |   | 3 | 557 | 957 | 949 | -0.014 | -0.311 | 0.129 | -0.023 | 0.033 |
|---|
| MICAL2 |   | 4 | 440 | 1659 | 1629 | -0.012 | 0.143 | -0.042 | -0.145 | 0.161 |
|---|
| ZC3H11A |   | 1 | 1195 | 1659 | 1678 | 0.035 | 0.256 | 0.097 | 0.191 | 0.079 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| FZD3 |   | 1 | 1195 | 1659 | 1678 | -0.099 | 0.180 | 0.125 | -0.018 | -0.126 |
|---|
| CALM1 |   | 6 | 301 | 614 | 602 | 0.014 | 0.021 | 0.192 | 0.221 | 0.247 |
|---|
| BTK |   | 2 | 743 | 1492 | 1474 | -0.034 | -0.210 | -0.081 | -0.218 | -0.046 |
|---|
| FHL2 |   | 21 | 73 | 658 | 630 | 0.143 | 0.239 | -0.033 | 0.107 | 0.294 |
|---|
| CUX1 |   | 6 | 301 | 1203 | 1169 | -0.042 | 0.256 | -0.121 | 0.105 | 0.040 |
|---|
| PFKFB3 |   | 3 | 557 | 1659 | 1632 | 0.301 | 0.085 | 0.328 | 0.056 | 0.331 |
|---|
| SLC9A5 |   | 1 | 1195 | 1659 | 1678 | -0.030 | 0.222 | 0.074 | 0.171 | 0.087 |
|---|
| ADRBK2 |   | 1 | 1195 | 1659 | 1678 | -0.131 | -0.107 | 0.209 | -0.263 | -0.094 |
|---|
| PDHB |   | 13 | 124 | 525 | 503 | 0.056 | 0.017 | 0.197 | -0.205 | 0.193 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| WDR46 |   | 2 | 743 | 366 | 395 | 0.055 | 0.154 | 0.106 | -0.094 | 0.034 |
|---|
| DCLK1 |   | 7 | 256 | 141 | 151 | -0.145 | -0.006 | -0.048 | 0.188 | -0.035 |
|---|
| ARHGEF6 |   | 10 | 167 | 525 | 507 | -0.068 | -0.120 | 0.166 | undef | -0.115 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| ITPR1 |   | 4 | 440 | 1576 | 1543 | -0.096 | 0.077 | 0.015 | undef | 0.008 |
|---|
| IGFBP5 |   | 48 | 23 | 83 | 80 | -0.076 | 0.110 | 0.074 | 0.278 | 0.181 |
|---|
| NEK9 |   | 2 | 743 | 1659 | 1643 | -0.074 | 0.037 | 0.211 | undef | 0.056 |
|---|
| PAM |   | 10 | 167 | 638 | 621 | -0.035 | 0.307 | 0.200 | 0.002 | 0.104 |
|---|
| EIF2B2 |   | 1 | 1195 | 1659 | 1678 | 0.082 | -0.003 | 0.081 | 0.101 | 0.214 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| FHL1 |   | 14 | 117 | 614 | 591 | -0.041 | 0.104 | 0.301 | undef | 0.266 |
|---|
| ARRB2 |   | 2 | 743 | 1396 | 1391 | -0.064 | -0.239 | -0.017 | undef | -0.189 |
|---|
| SNCAIP |   | 10 | 167 | 366 | 362 | -0.091 | 0.013 | 0.092 | 0.136 | 0.059 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
| TRIM29 |   | 8 | 222 | 296 | 287 | 0.161 | 0.240 | -0.150 | -0.077 | 0.024 |
|---|
GO Enrichment output for subnetwork 6561 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair. DNA ligation | GO:0006288 |  | 3.425E-12 | 8.366E-09 |
|---|
| DNA ligation during DNA repair | GO:0051103 |  | 7.132E-11 | 8.712E-08 |
|---|
| regulation of transcriptional preinitiation complex assembly | GO:0045898 |  | 7.132E-11 | 5.808E-08 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 7.754E-11 | 4.736E-08 |
|---|
| regulation of transcription initiation from RNA polymerase II promoter | GO:0060260 |  | 4.243E-10 | 2.073E-07 |
|---|
| DNA ligation | GO:0006266 |  | 8.452E-10 | 3.441E-07 |
|---|
| base-excision repair | GO:0006284 |  | 5.882E-09 | 2.053E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.015E-08 | 6.155E-06 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 2.015E-08 | 5.471E-06 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 4.987E-08 | 1.218E-05 |
|---|
| ear development | GO:0043583 |  | 5.151E-08 | 1.144E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| base-excision repair. DNA ligation | GO:0006288 |  | 5.243E-12 | 1.261E-08 |
|---|
| DNA ligation during DNA repair | GO:0051103 |  | 1.091E-10 | 1.313E-07 |
|---|
| regulation of transcriptional preinitiation complex assembly | GO:0045898 |  | 1.091E-10 | 8.75E-08 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.503E-10 | 9.041E-08 |
|---|
| regulation of transcription initiation from RNA polymerase II promoter | GO:0060260 |  | 6.487E-10 | 3.122E-07 |
|---|
| DNA ligation | GO:0006266 |  | 1.292E-09 | 5.179E-07 |
|---|
| base-excision repair | GO:0006284 |  | 7.667E-09 | 2.635E-06 |
|---|
| auditory receptor cell differentiation | GO:0042491 |  | 2.178E-08 | 6.552E-06 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 3.072E-08 | 8.213E-06 |
|---|
| ear development | GO:0043583 |  | 5.866E-08 | 1.411E-05 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 7.593E-08 | 1.661E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.329E-10 | 5.357E-07 |
|---|
| DNA ligation during DNA repair | GO:0051103 |  | 3.029E-08 | 3.484E-05 |
|---|
| regulation of transcriptional preinitiation complex assembly | GO:0045898 |  | 3.029E-08 | 2.323E-05 |
|---|
| regulation of transcription initiation from RNA polymerase II promoter | GO:0060260 |  | 1.399E-07 | 8.044E-05 |
|---|
| DNA ligation | GO:0006266 |  | 2.505E-07 | 1.152E-04 |
|---|
| base-excision repair | GO:0006284 |  | 5.147E-07 | 1.973E-04 |
|---|
| response to UV | GO:0009411 |  | 6.572E-07 | 2.159E-04 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.938E-06 | 5.572E-04 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.155E-06 | 5.508E-04 |
|---|
| microvillus organization | GO:0032528 |  | 3.045E-06 | 7.003E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 4.536E-06 | 9.484E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.5E-06 | 2.765E-03 |
|---|
| meiosis I | GO:0007127 |  | 2.291E-06 | 2.111E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 2.291E-06 | 1.407E-03 |
|---|
| negative regulation of DNA metabolic process | GO:0051053 |  | 2.89E-06 | 1.332E-03 |
|---|
| somatic diversification of immune receptors via somatic mutation | GO:0002566 |  | 3.074E-06 | 1.133E-03 |
|---|
| ER overload response | GO:0006983 |  | 3.074E-06 | 9.444E-04 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 6.922E-06 | 1.823E-03 |
|---|
| immunoglobulin production during immune response | GO:0002381 |  | 8.543E-06 | 1.968E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.084E-05 | 2.221E-03 |
|---|
| somatic recombination of immunoglobulin gene segments | GO:0016447 |  | 1.277E-05 | 2.353E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.473E-05 | 2.469E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.329E-10 | 5.357E-07 |
|---|
| DNA ligation during DNA repair | GO:0051103 |  | 3.029E-08 | 3.484E-05 |
|---|
| regulation of transcriptional preinitiation complex assembly | GO:0045898 |  | 3.029E-08 | 2.323E-05 |
|---|
| regulation of transcription initiation from RNA polymerase II promoter | GO:0060260 |  | 1.399E-07 | 8.044E-05 |
|---|
| DNA ligation | GO:0006266 |  | 2.505E-07 | 1.152E-04 |
|---|
| base-excision repair | GO:0006284 |  | 5.147E-07 | 1.973E-04 |
|---|
| response to UV | GO:0009411 |  | 6.572E-07 | 2.159E-04 |
|---|
| DNA unwinding during replication | GO:0006268 |  | 1.938E-06 | 5.572E-04 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.155E-06 | 5.508E-04 |
|---|
| microvillus organization | GO:0032528 |  | 3.045E-06 | 7.003E-04 |
|---|
| DNA duplex unwinding | GO:0032508 |  | 4.536E-06 | 9.484E-04 |
|---|



