Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6533
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7549 | 3.450e-03 | 5.580e-04 | 7.680e-01 | 1.478e-06 |
|---|
| Loi | 0.2303 | 8.024e-02 | 1.135e-02 | 4.281e-01 | 3.900e-04 |
|---|
| Schmidt | 0.6728 | 0.000e+00 | 0.000e+00 | 4.558e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.475e-03 | 0.000e+00 |
|---|
| Wang | 0.2615 | 2.694e-03 | 4.396e-02 | 3.970e-01 | 4.703e-05 |
|---|
Expression data for subnetwork 6533 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
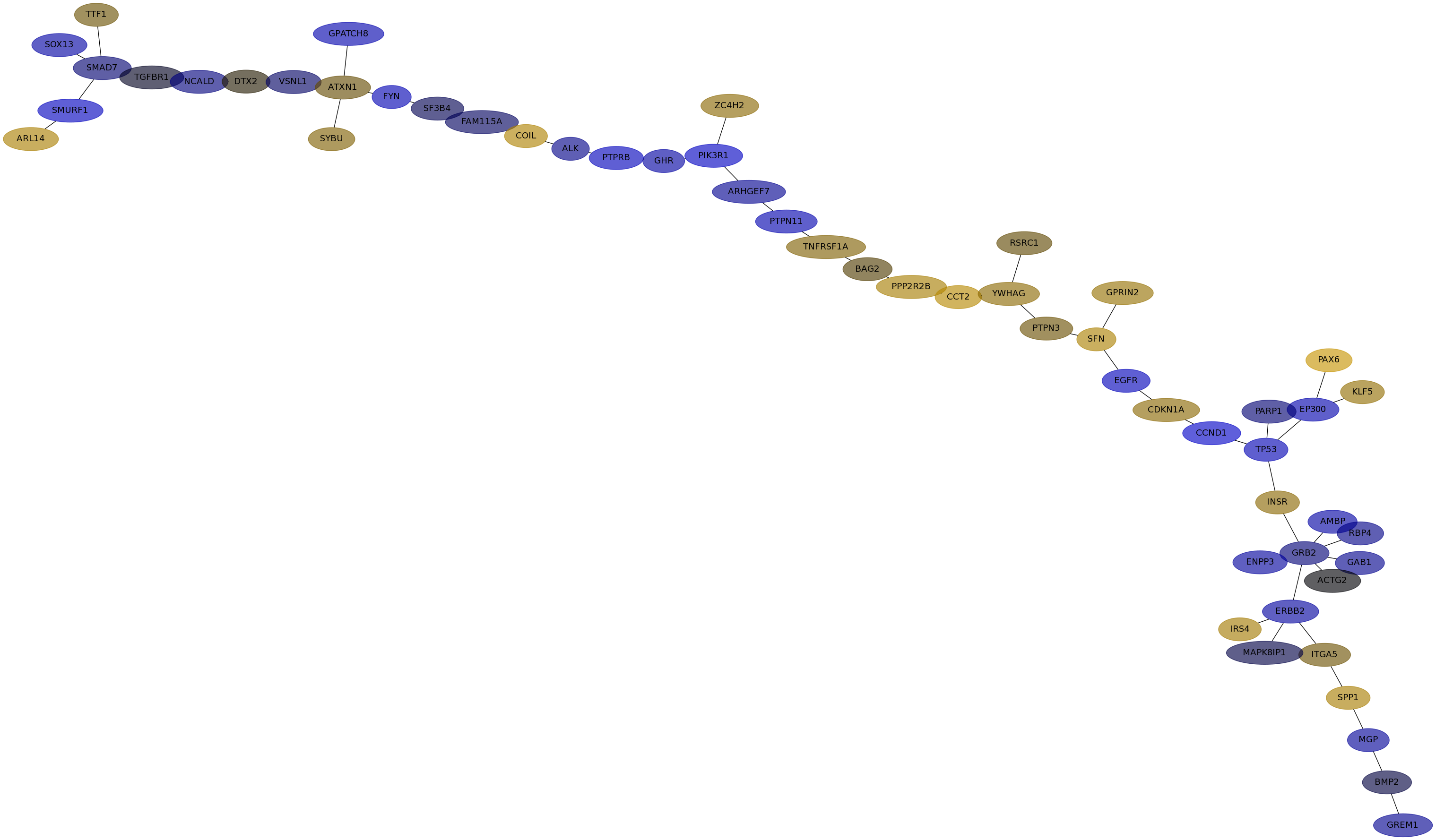
Score for each gene in subnetwork 6533 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| FAM115A |   | 1 | 1195 | 1793 | 1810 | -0.023 | 0.012 | -0.002 | 0.339 | 0.153 |
|---|
| PAX6 |   | 22 | 69 | 318 | 312 | 0.259 | -0.003 | 0.112 | undef | 0.168 |
|---|
| ARHGEF7 |   | 25 | 59 | 83 | 92 | -0.074 | -0.061 | 0.208 | 0.028 | 0.015 |
|---|
| MGP |   | 3 | 557 | 1450 | 1430 | -0.088 | 0.007 | 0.150 | -0.084 | -0.030 |
|---|
| SMAD7 |   | 5 | 360 | 1034 | 1004 | -0.035 | 0.131 | 0.169 | 0.061 | 0.147 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| TGFBR1 |   | 4 | 440 | 1793 | 1755 | -0.004 | 0.036 | -0.078 | 0.181 | undef |
|---|
| PTPN3 |   | 11 | 148 | 658 | 635 | 0.029 | 0.060 | -0.067 | 0.225 | -0.034 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| BAG2 |   | 4 | 440 | 1793 | 1755 | 0.016 | 0.178 | 0.152 | -0.249 | 0.083 |
|---|
| ACTG2 |   | 1 | 1195 | 1793 | 1810 | -0.001 | 0.095 | 0.232 | -0.035 | 0.121 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| TTF1 |   | 2 | 743 | 1793 | 1779 | 0.031 | 0.244 | 0.114 | 0.231 | 0.008 |
|---|
| BMP2 |   | 11 | 148 | 525 | 505 | -0.011 | 0.097 | 0.148 | -0.114 | -0.069 |
|---|
| CCND1 |   | 26 | 52 | 1 | 24 | -0.258 | 0.074 | 0.117 | undef | 0.003 |
|---|
| SPP1 |   | 26 | 52 | 83 | 91 | 0.130 | 0.119 | 0.184 | 0.204 | 0.153 |
|---|
| ARL14 |   | 1 | 1195 | 1793 | 1810 | 0.135 | 0.019 | -0.122 | undef | undef |
|---|
| PTPRB |   | 5 | 360 | 991 | 977 | -0.199 | 0.006 | 0.149 | 0.094 | 0.131 |
|---|
| AMBP |   | 1 | 1195 | 1793 | 1810 | -0.115 | 0.023 | -0.088 | 0.206 | 0.208 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| GPATCH8 |   | 8 | 222 | 614 | 599 | -0.143 | 0.014 | 0.019 | -0.070 | 0.061 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| PTPN11 |   | 20 | 81 | 83 | 94 | -0.152 | 0.014 | 0.066 | 0.304 | 0.089 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| ENPP3 |   | 1 | 1195 | 1793 | 1810 | -0.099 | 0.027 | -0.025 | 0.099 | 0.134 |
|---|
| GREM1 |   | 9 | 196 | 525 | 508 | -0.073 | 0.200 | 0.028 | 0.225 | 0.261 |
|---|
| CCT2 |   | 3 | 557 | 1474 | 1450 | 0.182 | 0.008 | 0.043 | 0.055 | 0.004 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| ZC4H2 |   | 17 | 95 | 570 | 546 | 0.065 | 0.061 | 0.304 | -0.072 | 0.059 |
|---|
| SYBU |   | 3 | 557 | 1055 | 1043 | 0.049 | 0.010 | -0.088 | 0.166 | 0.008 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| VSNL1 |   | 3 | 557 | 1437 | 1418 | -0.026 | 0.080 | 0.220 | -0.099 | -0.041 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| IRS4 |   | 32 | 40 | 236 | 224 | 0.115 | -0.111 | -0.088 | 0.096 | 0.142 |
|---|
| MAPK8IP1 |   | 4 | 440 | 1576 | 1543 | -0.012 | 0.058 | -0.020 | 0.282 | -0.049 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| SOX13 |   | 4 | 440 | 1034 | 1015 | -0.113 | 0.095 | -0.002 | 0.271 | -0.184 |
|---|
| ALK |   | 2 | 743 | 1793 | 1779 | -0.062 | 0.048 | 0.231 | 0.083 | -0.080 |
|---|
| GRB2 |   | 13 | 124 | 318 | 316 | -0.040 | -0.159 | -0.036 | -0.028 | -0.024 |
|---|
| RBP4 |   | 4 | 440 | 764 | 761 | -0.059 | -0.011 | 0.206 | 0.334 | -0.045 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| FYN |   | 10 | 167 | 1 | 40 | -0.171 | -0.167 | 0.073 | -0.137 | 0.110 |
|---|
| SF3B4 |   | 2 | 743 | 1351 | 1344 | -0.017 | 0.017 | -0.199 | 0.167 | -0.007 |
|---|
| NCALD |   | 4 | 440 | 1709 | 1672 | -0.046 | 0.078 | 0.090 | undef | 0.004 |
|---|
| TNFRSF1A |   | 8 | 222 | 412 | 413 | 0.050 | 0.094 | -0.014 | 0.214 | -0.045 |
|---|
| DTX2 |   | 2 | 743 | 1793 | 1779 | 0.005 | 0.174 | -0.122 | undef | -0.200 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| SMURF1 |   | 5 | 360 | 682 | 671 | -0.222 | 0.236 | -0.049 | undef | -0.006 |
|---|
| GAB1 |   | 5 | 360 | 1102 | 1076 | -0.069 | 0.084 | -0.117 | 0.116 | -0.212 |
|---|
| COIL |   | 4 | 440 | 913 | 904 | 0.151 | 0.144 | 0.071 | 0.106 | 0.044 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6533 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 1.954E-12 | 4.773E-09 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 1.053E-09 | 1.287E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 4.3E-09 | 3.502E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 8.709E-09 | 5.319E-06 |
|---|
| organ growth | GO:0035265 |  | 8.709E-09 | 4.255E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 8.709E-09 | 3.546E-06 |
|---|
| cellular response to insulin stimulus | GO:0032869 |  | 9.473E-09 | 3.306E-06 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 1.092E-08 | 3.334E-06 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 1.116E-08 | 3.029E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.736E-08 | 4.242E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 3.116E-08 | 6.92E-06 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 7.152E-11 | 1.721E-07 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 6.036E-09 | 7.261E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 1.045E-08 | 8.382E-06 |
|---|
| organ growth | GO:0035265 |  | 1.045E-08 | 6.286E-06 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 1.045E-08 | 5.029E-06 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 1.234E-08 | 4.947E-06 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 1.366E-08 | 4.695E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 2.083E-08 | 6.266E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 3.738E-08 | 9.992E-06 |
|---|
| regulation of insulin secretion | GO:0050796 |  | 6.116E-08 | 1.472E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 7.547E-08 | 1.651E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 8.407E-11 | 1.934E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 2.593E-09 | 2.981E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.749E-09 | 5.941E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 7.924E-09 | 4.557E-06 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 1.101E-08 | 5.065E-06 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 1.426E-08 | 5.465E-06 |
|---|
| organ growth | GO:0035265 |  | 1.801E-08 | 5.919E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.801E-08 | 5.179E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 3.589E-08 | 9.173E-06 |
|---|
| regulation of insulin secretion | GO:0050796 |  | 4.617E-08 | 1.062E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.534E-08 | 1.993E-05 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 9.114E-08 | 1.68E-04 |
|---|
| regulation of MAP kinase activity | GO:0043405 |  | 5.591E-07 | 5.152E-04 |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 2.206E-06 | 1.355E-03 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 2.704E-06 | 1.246E-03 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 3.095E-06 | 1.141E-03 |
|---|
| positive regulation of growth | GO:0045927 |  | 3.643E-06 | 1.119E-03 |
|---|
| glucose homeostasis | GO:0042593 |  | 4.357E-06 | 1.147E-03 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 5.54E-06 | 1.276E-03 |
|---|
| response to hormone stimulus | GO:0009725 |  | 6.537E-06 | 1.339E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 8.283E-06 | 1.527E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.179E-05 | 1.976E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cellular response to hormone stimulus | GO:0032870 |  | 8.407E-11 | 1.934E-07 |
|---|
| negative regulation of peptide secretion | GO:0002792 |  | 2.593E-09 | 2.981E-06 |
|---|
| regulation of protein export from nucleus | GO:0046825 |  | 7.749E-09 | 5.941E-06 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 7.924E-09 | 4.557E-06 |
|---|
| carbohydrate homeostasis | GO:0033500 |  | 1.101E-08 | 5.065E-06 |
|---|
| positive regulation of hormone secretion | GO:0046887 |  | 1.426E-08 | 5.465E-06 |
|---|
| organ growth | GO:0035265 |  | 1.801E-08 | 5.919E-06 |
|---|
| regulation of growth hormone secretion | GO:0060123 |  | 1.801E-08 | 5.179E-06 |
|---|
| nerve growth factor receptor signaling pathway | GO:0048011 |  | 3.589E-08 | 9.173E-06 |
|---|
| regulation of insulin secretion | GO:0050796 |  | 4.617E-08 | 1.062E-05 |
|---|
| insulin receptor signaling pathway | GO:0008286 |  | 9.534E-08 | 1.993E-05 |
|---|



