Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6510
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7551 | 3.436e-03 | 5.550e-04 | 7.674e-01 | 1.463e-06 |
|---|
| Loi | 0.2302 | 8.041e-02 | 1.140e-02 | 4.286e-01 | 3.927e-04 |
|---|
| Schmidt | 0.6725 | 0.000e+00 | 0.000e+00 | 4.590e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.473e-03 | 0.000e+00 |
|---|
| Wang | 0.2613 | 2.715e-03 | 4.415e-02 | 3.979e-01 | 4.770e-05 |
|---|
Expression data for subnetwork 6510 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
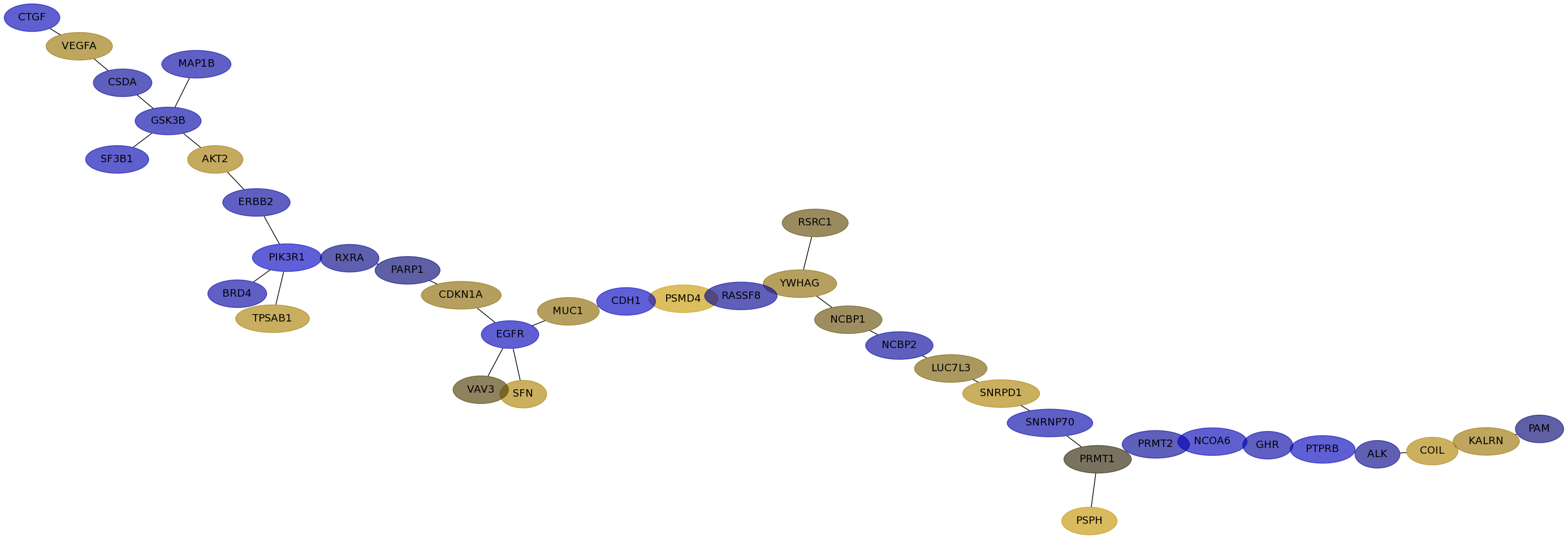
Score for each gene in subnetwork 6510 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| SF3B1 |   | 18 | 86 | 296 | 280 | -0.154 | 0.126 | -0.073 | -0.131 | -0.204 |
|---|
| RXRA |   | 7 | 256 | 457 | 451 | -0.055 | 0.134 | -0.027 | 0.022 | -0.096 |
|---|
| PRMT1 |   | 2 | 743 | 1203 | 1206 | 0.006 | -0.136 | 0.075 | -0.144 | -0.165 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| RASSF8 |   | 5 | 360 | 296 | 310 | -0.078 | 0.121 | 0.174 | 0.114 | -0.092 |
|---|
| SNRPD1 |   | 2 | 743 | 412 | 439 | 0.140 | -0.211 | 0.198 | -0.045 | -0.008 |
|---|
| PTPRB |   | 5 | 360 | 991 | 977 | -0.199 | 0.006 | 0.149 | 0.094 | 0.131 |
|---|
| CTGF |   | 32 | 40 | 318 | 290 | -0.169 | 0.181 | 0.126 | 0.101 | 0.209 |
|---|
| NCOA6 |   | 3 | 557 | 957 | 949 | -0.179 | -0.123 | -0.167 | 0.103 | 0.133 |
|---|
| CDH1 |   | 37 | 32 | 296 | 278 | -0.227 | 0.187 | -0.236 | 0.137 | -0.024 |
|---|
| GHR |   | 35 | 34 | 83 | 83 | -0.112 | 0.174 | 0.090 | 0.176 | 0.201 |
|---|
| CSDA |   | 1 | 1195 | 1840 | 1853 | -0.091 | 0.025 | 0.159 | -0.014 | 0.097 |
|---|
| VAV3 |   | 9 | 196 | 638 | 622 | 0.015 | 0.071 | -0.245 | 0.148 | 0.038 |
|---|
| BRD4 |   | 11 | 148 | 1047 | 1003 | -0.119 | 0.095 | -0.075 | 0.081 | -0.044 |
|---|
| PARP1 |   | 29 | 45 | 83 | 84 | -0.036 | -0.162 | -0.089 | 0.238 | 0.012 |
|---|
| AKT2 |   | 28 | 47 | 412 | 394 | 0.110 | -0.118 | -0.003 | 0.088 | -0.092 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| KALRN |   | 4 | 440 | 1102 | 1080 | 0.090 | 0.136 | -0.057 | 0.202 | -0.053 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| PSMD4 |   | 11 | 148 | 296 | 282 | 0.285 | -0.243 | -0.043 | 0.087 | -0.045 |
|---|
| PRMT2 |   | 5 | 360 | 957 | 938 | -0.081 | 0.046 | 0.089 | 0.235 | -0.128 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| ALK |   | 2 | 743 | 1793 | 1779 | -0.062 | 0.048 | 0.231 | 0.083 | -0.080 |
|---|
| MUC1 |   | 13 | 124 | 1547 | 1490 | 0.063 | 0.214 | -0.190 | undef | -0.132 |
|---|
| NCBP1 |   | 3 | 557 | 1294 | 1276 | 0.027 | -0.115 | 0.006 | -0.024 | -0.093 |
|---|
| LUC7L3 |   | 16 | 104 | 296 | 281 | 0.043 | 0.237 | 0.175 | 0.135 | 0.074 |
|---|
| PAM |   | 10 | 167 | 638 | 621 | -0.035 | 0.307 | 0.200 | 0.002 | 0.104 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| NCBP2 |   | 3 | 557 | 614 | 623 | -0.087 | 0.170 | 0.311 | -0.055 | -0.052 |
|---|
| TPSAB1 |   | 1 | 1195 | 1840 | 1853 | 0.131 | -0.181 | -0.002 | -0.035 | -0.087 |
|---|
| SNRNP70 |   | 5 | 360 | 296 | 310 | -0.129 | 0.130 | -0.059 | -0.099 | 0.036 |
|---|
| COIL |   | 4 | 440 | 913 | 904 | 0.151 | 0.144 | 0.071 | 0.106 | 0.044 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
| PSPH |   | 20 | 81 | 366 | 356 | 0.240 | -0.019 | -0.102 | 0.358 | 0.025 |
|---|
GO Enrichment output for subnetwork 6510 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 3.526E-09 | 8.615E-06 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 5.961E-09 | 7.281E-06 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.193E-08 | 1.785E-05 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 2.716E-07 | 1.659E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 2.928E-07 | 1.431E-04 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 5.42E-07 | 2.207E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 8.71E-07 | 3.04E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 1.511E-06 | 4.614E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 2.261E-06 | 6.138E-04 |
|---|
| L-serine metabolic process | GO:0006563 |  | 4.422E-06 | 1.08E-03 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 9.691E-06 | 2.152E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 5.717E-09 | 1.375E-05 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 9.659E-09 | 1.162E-05 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.922E-08 | 2.343E-05 |
|---|
| L-serine biosynthetic process | GO:0006564 |  | 3.643E-07 | 2.191E-04 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 3.643E-07 | 1.753E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 4.717E-07 | 1.892E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.399E-06 | 4.81E-04 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 2.025E-06 | 6.091E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 3.03E-06 | 8.101E-04 |
|---|
| L-serine metabolic process | GO:0006563 |  | 4.318E-06 | 1.039E-03 |
|---|
| serine family amino acid biosynthetic process | GO:0009070 |  | 1.022E-05 | 2.235E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 8.627E-09 | 1.984E-05 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.61E-08 | 1.851E-05 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 3.609E-08 | 2.767E-05 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 8.164E-07 | 4.694E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.088E-06 | 5.006E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 2.382E-06 | 9.13E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.528E-06 | 1.488E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.528E-06 | 1.302E-03 |
|---|
| phosphatidylinositol metabolic process | GO:0046488 |  | 5.344E-05 | 0.01365669 |
|---|
| positive regulation of growth | GO:0045927 |  | 6.87E-05 | 0.01580089 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.43E-05 | 0.0155351 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 1.414E-07 | 2.606E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 7.667E-07 | 7.065E-04 |
|---|
| positive regulation of growth | GO:0045927 |  | 3.643E-06 | 2.238E-03 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 7.124E-06 | 3.282E-03 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 1.26E-05 | 4.644E-03 |
|---|
| protein amino acid methylation | GO:0006479 |  | 1.616E-05 | 4.965E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 3.531E-05 | 9.297E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 7.984E-05 | 0.01839374 |
|---|
| biopolymer methylation | GO:0043414 |  | 1.455E-04 | 0.02980142 |
|---|
| regulation of intracellular protein transport | GO:0033157 |  | 2.152E-04 | 0.03966668 |
|---|
| mRNA stabilization | GO:0048255 |  | 2.211E-04 | 0.03704786 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of nucleocytoplasmic transport | GO:0046824 |  | 8.627E-09 | 1.984E-05 |
|---|
| positive regulation of intracellular transport | GO:0032388 |  | 1.61E-08 | 1.851E-05 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 3.609E-08 | 2.767E-05 |
|---|
| regulation of nucleobase. nucleoside. nucleotide and nucleic acid transport | GO:0032239 |  | 8.164E-07 | 4.694E-04 |
|---|
| regulation of nucleocytoplasmic transport | GO:0046822 |  | 1.088E-06 | 5.006E-04 |
|---|
| regulation of intracellular transport | GO:0032386 |  | 2.382E-06 | 9.13E-04 |
|---|
| insulin-like growth factor receptor signaling pathway | GO:0048009 |  | 4.528E-06 | 1.488E-03 |
|---|
| phosphoinositide 3-kinase cascade | GO:0014065 |  | 4.528E-06 | 1.302E-03 |
|---|
| phosphatidylinositol metabolic process | GO:0046488 |  | 5.344E-05 | 0.01365669 |
|---|
| positive regulation of growth | GO:0045927 |  | 6.87E-05 | 0.01580089 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.43E-05 | 0.0155351 |
|---|



