Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6505
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7552 | 3.433e-03 | 5.550e-04 | 7.673e-01 | 1.462e-06 |
|---|
| Loi | 0.2302 | 8.042e-02 | 1.140e-02 | 4.286e-01 | 3.929e-04 |
|---|
| Schmidt | 0.6725 | 0.000e+00 | 0.000e+00 | 4.587e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7907 | 0.000e+00 | 0.000e+00 | 3.476e-03 | 0.000e+00 |
|---|
| Wang | 0.2613 | 2.715e-03 | 4.415e-02 | 3.979e-01 | 4.769e-05 |
|---|
Expression data for subnetwork 6505 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
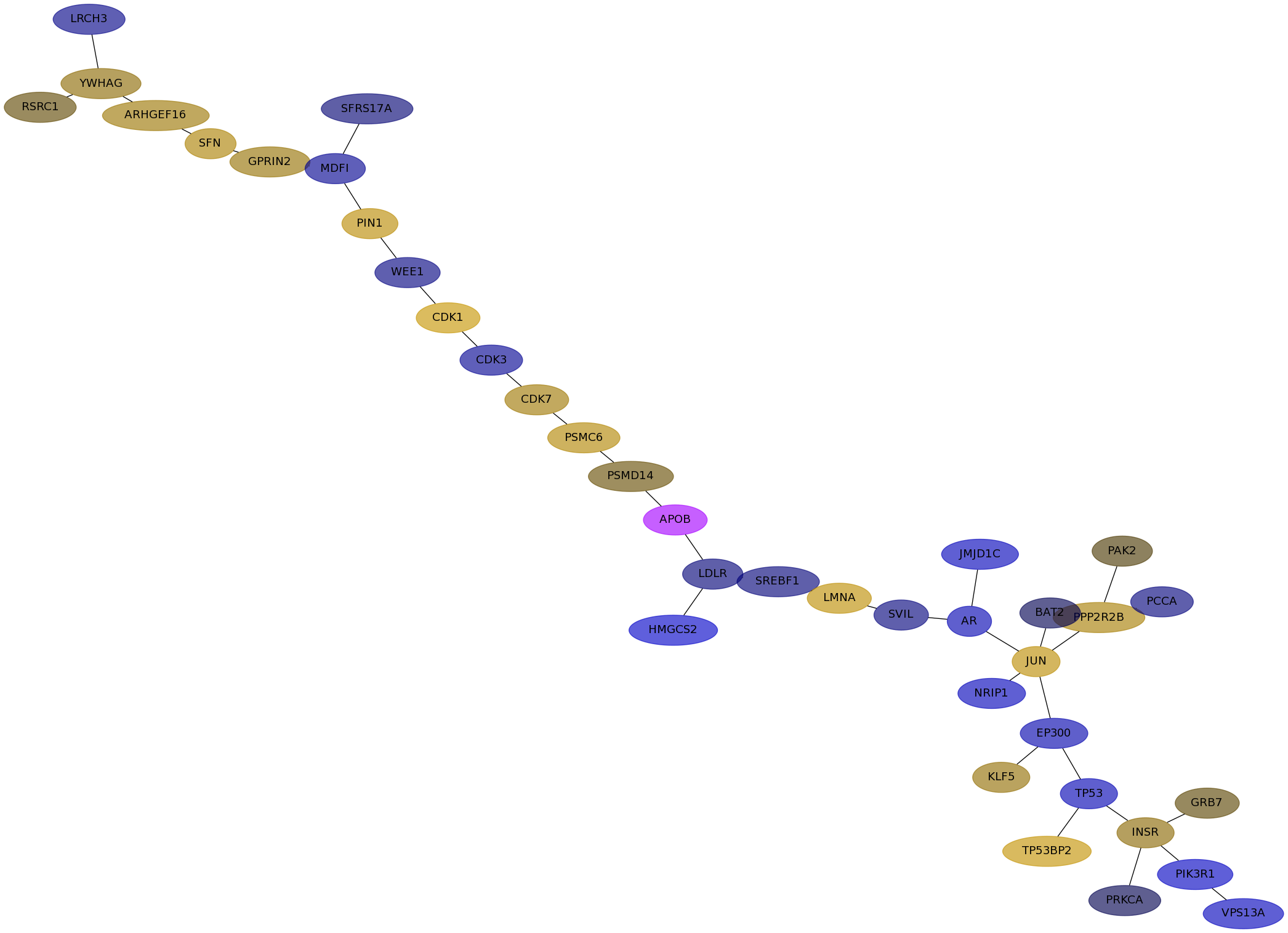
Score for each gene in subnetwork 6505 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| LDLR |   | 5 | 360 | 780 | 767 | -0.041 | 0.218 | 0.039 | -0.028 | 0.064 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| SFRS17A |   | 3 | 557 | 913 | 908 | -0.036 | 0.169 | -0.225 | -0.176 | -0.078 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| VPS13A |   | 10 | 167 | 236 | 234 | -0.203 | 0.105 | -0.046 | 0.120 | 0.062 |
|---|
| APOB |   | 3 | 557 | 1771 | 1745 | undef | 0.001 | 0.156 | 0.151 | 0.144 |
|---|
| RSRC1 |   | 21 | 73 | 366 | 354 | 0.023 | 0.104 | 0.105 | -0.098 | 0.212 |
|---|
| LMNA |   | 3 | 557 | 1047 | 1038 | 0.208 | 0.223 | -0.017 | 0.247 | 0.069 |
|---|
| ARHGEF16 |   | 10 | 167 | 296 | 283 | 0.096 | 0.036 | -0.047 | 0.200 | 0.038 |
|---|
| CDK3 |   | 7 | 256 | 590 | 570 | -0.076 | 0.189 | -0.041 | -0.089 | 0.082 |
|---|
| JUN |   | 26 | 52 | 179 | 177 | 0.200 | 0.051 | 0.220 | undef | 0.200 |
|---|
| WEE1 |   | 4 | 440 | 658 | 649 | -0.051 | 0.163 | 0.034 | undef | 0.215 |
|---|
| GPRIN2 |   | 16 | 104 | 179 | 190 | 0.084 | 0.078 | 0.012 | 0.054 | 0.114 |
|---|
| PIN1 |   | 24 | 62 | 179 | 181 | 0.194 | 0.040 | -0.040 | 0.065 | -0.048 |
|---|
| PAK2 |   | 26 | 52 | 179 | 177 | 0.014 | 0.161 | 0.081 | -0.044 | 0.062 |
|---|
| SREBF1 |   | 9 | 196 | 478 | 460 | -0.042 | 0.232 | -0.195 | 0.140 | -0.074 |
|---|
| PSMD14 |   | 4 | 440 | 842 | 831 | 0.027 | 0.135 | -0.179 | -0.018 | -0.031 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| TP53BP2 |   | 11 | 148 | 692 | 668 | 0.245 | -0.012 | 0.129 | 0.021 | 0.017 |
|---|
| NRIP1 |   | 1 | 1195 | 1825 | 1838 | -0.193 | 0.042 | -0.054 | 0.154 | 0.008 |
|---|
| LRCH3 |   | 1 | 1195 | 1825 | 1838 | -0.059 | 0.229 | 0.162 | -0.057 | 0.225 |
|---|
| MDFI |   | 17 | 95 | 236 | 226 | -0.075 | -0.001 | 0.177 | 0.169 | -0.112 |
|---|
| BAT2 |   | 3 | 557 | 957 | 949 | -0.017 | 0.031 | 0.058 | undef | 0.016 |
|---|
| PCCA |   | 2 | 743 | 1825 | 1808 | -0.045 | 0.042 | 0.100 | 0.145 | -0.016 |
|---|
| JMJD1C |   | 7 | 256 | 552 | 542 | -0.195 | 0.241 | 0.074 | undef | 0.129 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| HMGCS2 |   | 2 | 743 | 1825 | 1808 | -0.277 | 0.226 | -0.194 | 0.151 | -0.085 |
|---|
| CDK7 |   | 7 | 256 | 318 | 328 | 0.105 | 0.184 | 0.017 | 0.078 | -0.097 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| YWHAG |   | 24 | 62 | 296 | 279 | 0.067 | 0.024 | undef | 0.266 | undef |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| PSMC6 |   | 5 | 360 | 318 | 334 | 0.163 | 0.061 | -0.010 | 0.069 | 0.044 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| SVIL |   | 1 | 1195 | 1825 | 1838 | -0.045 | 0.135 | 0.080 | undef | -0.013 |
|---|
| AR |   | 46 | 24 | 141 | 133 | -0.163 | 0.081 | -0.035 | 0.178 | -0.030 |
|---|
| KLF5 |   | 76 | 9 | 1 | 7 | 0.077 | 0.117 | -0.007 | -0.110 | -0.006 |
|---|
| SFN |   | 25 | 59 | 478 | 454 | 0.141 | 0.270 | 0.023 | -0.102 | 0.101 |
|---|
GO Enrichment output for subnetwork 6505 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| melatonin biosynthetic process | GO:0030187 |  | 3.727E-10 | 9.105E-07 |
|---|
| indolalkylamine biosynthetic process | GO:0046219 |  | 1.115E-09 | 1.363E-06 |
|---|
| indolalkylamine metabolic process | GO:0006586 |  | 5.232E-08 | 4.261E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 6.275E-08 | 3.832E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 6.846E-08 | 3.345E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 7.309E-08 | 2.976E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 9.58E-08 | 3.344E-05 |
|---|
| cholesterol metabolic process | GO:0008203 |  | 2.203E-07 | 6.726E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.383E-07 | 6.468E-05 |
|---|
| lipoprotein particle clearance | GO:0034381 |  | 2.567E-07 | 6.27E-05 |
|---|
| lipoprotein catabolic process | GO:0042159 |  | 2.567E-07 | 5.7E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| indolalkylamine biosynthetic process | GO:0046219 |  | 3.719E-10 | 8.948E-07 |
|---|
| indolalkylamine metabolic process | GO:0006586 |  | 2.421E-08 | 2.913E-05 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 5.184E-08 | 4.157E-05 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 5.672E-08 | 3.411E-05 |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 7.294E-08 | 3.51E-05 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 8.019E-08 | 3.216E-05 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.368E-07 | 8.138E-05 |
|---|
| lipoprotein particle clearance | GO:0034381 |  | 2.569E-07 | 7.728E-05 |
|---|
| lipoprotein catabolic process | GO:0042159 |  | 2.569E-07 | 6.869E-05 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 7.568E-07 | 1.821E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 8.954E-07 | 1.959E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 1.373E-07 | 3.157E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.213E-07 | 4.845E-04 |
|---|
| lipoprotein particle clearance | GO:0034381 |  | 5.351E-07 | 4.103E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.651E-06 | 9.496E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.863E-06 | 8.568E-04 |
|---|
| negative regulation of cellular protein metabolic process | GO:0032269 |  | 2.137E-06 | 8.191E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.346E-06 | 7.707E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.55E-06 | 7.33E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 2.764E-06 | 7.064E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.782E-06 | 6.398E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.972E-06 | 6.214E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 2.65E-06 | 4.884E-03 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 5.651E-06 | 5.207E-03 |
|---|
| cellular response to starvation | GO:0009267 |  | 7.75E-06 | 4.761E-03 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 9.727E-06 | 4.482E-03 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 1.084E-05 | 3.997E-03 |
|---|
| negative regulation of transferase activity | GO:0051348 |  | 1.336E-05 | 4.103E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 1.337E-05 | 3.519E-03 |
|---|
| anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process | GO:0031145 |  | 1.627E-05 | 3.749E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity during mitotic cell cycle | GO:0051437 |  | 1.964E-05 | 4.021E-03 |
|---|
| response to starvation | GO:0042594 |  | 2.116E-05 | 3.899E-03 |
|---|
| positive regulation of ubiquitin-protein ligase activity | GO:0051443 |  | 2.15E-05 | 3.602E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein amino acid phosphorylation | GO:0001933 |  | 1.373E-07 | 3.157E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 4.213E-07 | 4.845E-04 |
|---|
| lipoprotein particle clearance | GO:0034381 |  | 5.351E-07 | 4.103E-04 |
|---|
| negative regulation of phosphorylation | GO:0042326 |  | 1.651E-06 | 9.496E-04 |
|---|
| negative regulation of insulin receptor signaling pathway | GO:0046627 |  | 1.863E-06 | 8.568E-04 |
|---|
| negative regulation of cellular protein metabolic process | GO:0032269 |  | 2.137E-06 | 8.191E-04 |
|---|
| negative regulation of phosphorus metabolic process | GO:0010563 |  | 2.346E-06 | 7.707E-04 |
|---|
| negative regulation of protein kinase activity | GO:0006469 |  | 2.55E-06 | 7.33E-04 |
|---|
| negative regulation of protein modification process | GO:0031400 |  | 2.764E-06 | 7.064E-04 |
|---|
| negative regulation of kinase activity | GO:0033673 |  | 2.782E-06 | 6.398E-04 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.972E-06 | 6.214E-04 |
|---|



