Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6449
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7556 | 3.405e-03 | 5.490e-04 | 7.660e-01 | 1.432e-06 |
|---|
| Loi | 0.2298 | 8.100e-02 | 1.155e-02 | 4.300e-01 | 4.024e-04 |
|---|
| Schmidt | 0.6726 | 0.000e+00 | 0.000e+00 | 4.584e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7906 | 0.000e+00 | 0.000e+00 | 3.486e-03 | 0.000e+00 |
|---|
| Wang | 0.2609 | 2.769e-03 | 4.464e-02 | 4.002e-01 | 4.947e-05 |
|---|
Expression data for subnetwork 6449 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
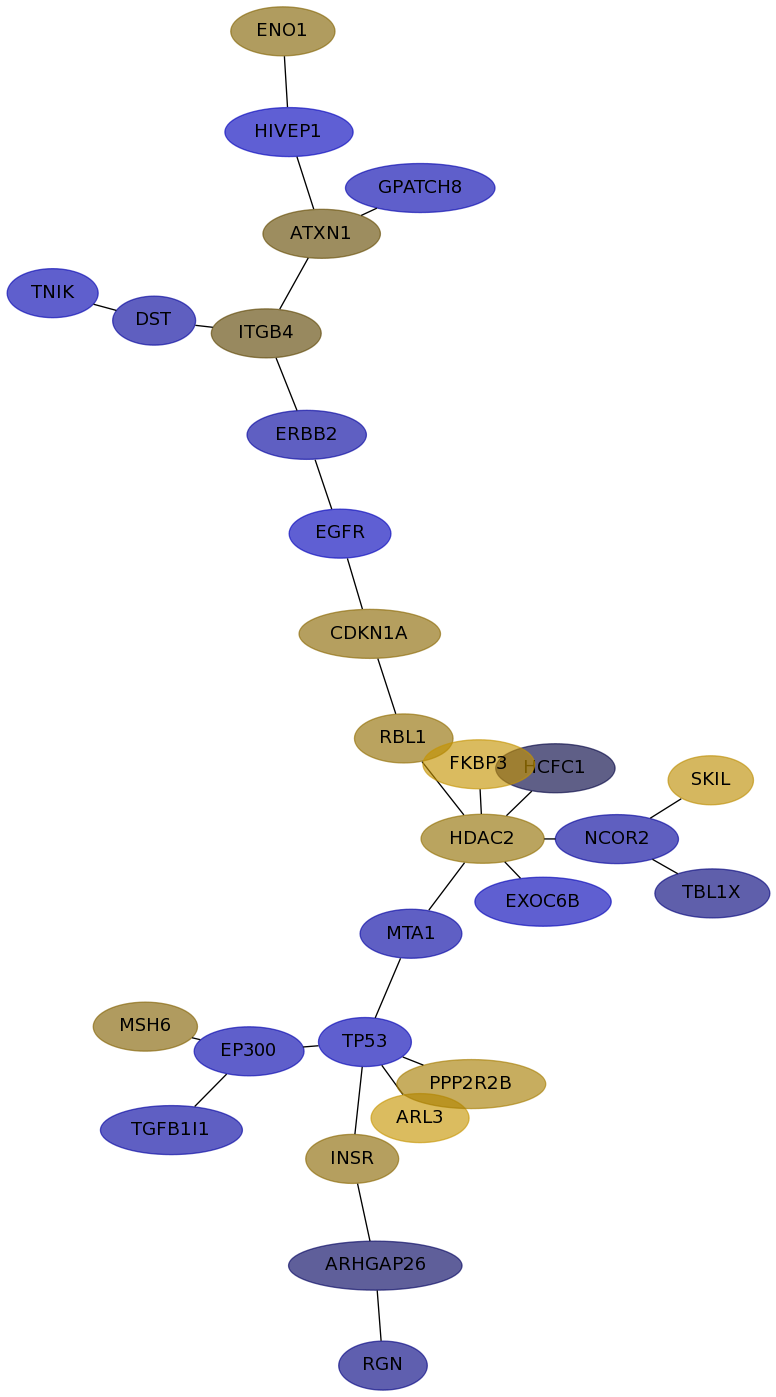
Score for each gene in subnetwork 6449 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| HCFC1 |   | 4 | 440 | 648 | 645 | -0.011 | 0.009 | 0.209 | 0.061 | -0.093 |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| ARHGAP26 |   | 21 | 73 | 457 | 438 | -0.023 | 0.171 | -0.222 | 0.015 | -0.348 |
|---|
| TNIK |   | 3 | 557 | 614 | 623 | -0.156 | 0.084 | undef | 0.072 | -0.090 |
|---|
| NCOR2 |   | 32 | 40 | 318 | 290 | -0.096 | 0.211 | -0.163 | 0.158 | 0.114 |
|---|
| EXOC6B |   | 6 | 301 | 318 | 330 | -0.183 | 0.205 | -0.104 | 0.030 | 0.037 |
|---|
| TP53 |   | 85 | 7 | 1 | 6 | -0.167 | 0.148 | -0.027 | 0.252 | 0.147 |
|---|
| ATXN1 |   | 17 | 95 | 1 | 29 | 0.026 | 0.192 | 0.175 | 0.125 | 0.163 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| FKBP3 |   | 2 | 743 | 2038 | 2020 | 0.255 | 0.091 | 0.062 | 0.299 | -0.068 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| RGN |   | 6 | 301 | 570 | 557 | -0.051 | -0.050 | 0.184 | 0.139 | -0.120 |
|---|
| ARL3 |   | 6 | 301 | 83 | 105 | 0.266 | -0.002 | 0.203 | 0.206 | -0.029 |
|---|
| DST |   | 22 | 69 | 1 | 25 | -0.094 | 0.343 | 0.065 | -0.062 | 0.194 |
|---|
| SKIL |   | 3 | 557 | 780 | 781 | 0.209 | 0.108 | 0.073 | 0.094 | -0.020 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| RBL1 |   | 21 | 73 | 1 | 27 | 0.078 | undef | undef | 0.051 | undef |
|---|
| MTA1 |   | 2 | 743 | 2038 | 2020 | -0.112 | 0.103 | -0.107 | 0.155 | -0.048 |
|---|
| GPATCH8 |   | 8 | 222 | 614 | 599 | -0.143 | 0.014 | 0.019 | -0.070 | 0.061 |
|---|
| TBL1X |   | 4 | 440 | 1735 | 1699 | -0.044 | 0.017 | -0.192 | 0.198 | -0.009 |
|---|
| PPP2R2B |   | 39 | 29 | 179 | 173 | 0.124 | -0.019 | 0.029 | 0.116 | -0.073 |
|---|
| ITGB4 |   | 35 | 34 | 1 | 20 | 0.021 | 0.313 | -0.134 | 0.080 | -0.114 |
|---|
| ENO1 |   | 3 | 557 | 1437 | 1418 | 0.054 | 0.211 | 0.010 | 0.017 | 0.178 |
|---|
| MSH6 |   | 12 | 140 | 1 | 36 | 0.052 | -0.057 | 0.051 | 0.126 | 0.064 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
| HIVEP1 |   | 6 | 301 | 764 | 758 | -0.199 | 0.069 | 0.000 | 0.193 | -0.042 |
|---|
| HDAC2 |   | 13 | 124 | 318 | 316 | 0.079 | -0.094 | 0.148 | 0.089 | 0.037 |
|---|
GO Enrichment output for subnetwork 6449 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 4.957E-07 | 1.211E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 7.501E-07 | 9.162E-04 |
|---|
| vesicle docking during exocytosis | GO:0006904 |  | 8.407E-07 | 6.846E-04 |
|---|
| vesicle docking | GO:0048278 |  | 1.146E-06 | 7.002E-04 |
|---|
| membrane docking | GO:0022406 |  | 2.888E-06 | 1.411E-03 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 8.042E-06 | 3.274E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.71E-05 | 5.967E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.77E-05 | 8.458E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 3.292E-05 | 8.935E-03 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 3.945E-05 | 9.638E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 4.189E-05 | 9.304E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 2.664E-07 | 6.409E-04 |
|---|
| regulation of helicase activity | GO:0051095 |  | 5.805E-07 | 6.984E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.721E-06 | 4.588E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.107E-05 | 6.659E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.818E-05 | 8.749E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.846E-05 | 7.401E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 2.484E-05 | 8.537E-03 |
|---|
| negative regulation of cell growth | GO:0030308 |  | 2.819E-05 | 8.477E-03 |
|---|
| negative regulation of cell size | GO:0045792 |  | 3.167E-05 | 8.466E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 3.252E-05 | 7.825E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 3.689E-05 | 8.069E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 4.943E-07 | 1.137E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 7.631E-07 | 8.775E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.044E-05 | 8.008E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.681E-05 | 9.667E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.934E-05 | 8.894E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.526E-05 | 9.681E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 3.581E-05 | 0.01176753 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.203E-05 | 0.01208406 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 4.892E-05 | 0.01250103 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 5.65E-05 | 0.01299559 |
|---|
| protein tetramerization | GO:0051262 |  | 5.65E-05 | 0.01181418 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 2.206E-06 | 4.065E-03 |
|---|
| response to UV | GO:0009411 |  | 2.553E-06 | 2.352E-03 |
|---|
| positive regulation of hydrolase activity | GO:0051345 |  | 3.971E-06 | 2.44E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 6.044E-06 | 2.785E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 1.043E-05 | 3.844E-03 |
|---|
| protein tetramerization | GO:0051262 |  | 1.043E-05 | 3.203E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 1.324E-05 | 3.486E-03 |
|---|
| positive regulation of MAP kinase activity | GO:0043406 |  | 2.173E-05 | 5.007E-03 |
|---|
| regulation of DNA metabolic process | GO:0051052 |  | 2.357E-05 | 4.826E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 2.758E-05 | 5.082E-03 |
|---|
| DNA damage response. signal transduction resulting in induction of apoptosis | GO:0008630 |  | 2.941E-05 | 4.928E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| response to UV | GO:0009411 |  | 4.943E-07 | 1.137E-03 |
|---|
| regulation of helicase activity | GO:0051095 |  | 7.631E-07 | 8.775E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.044E-05 | 8.008E-03 |
|---|
| protein amino acid autophosphorylation | GO:0046777 |  | 1.681E-05 | 9.667E-03 |
|---|
| response to light stimulus | GO:0009416 |  | 1.934E-05 | 8.894E-03 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 2.526E-05 | 9.681E-03 |
|---|
| positive regulation of epithelial cell proliferation | GO:0050679 |  | 3.581E-05 | 0.01176753 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 4.203E-05 | 0.01208406 |
|---|
| cellular response to extracellular stimulus | GO:0031668 |  | 4.892E-05 | 0.01250103 |
|---|
| regulation of fibroblast proliferation | GO:0048145 |  | 5.65E-05 | 0.01299559 |
|---|
| protein tetramerization | GO:0051262 |  | 5.65E-05 | 0.01181418 |
|---|



