Study Desmedt-ER-neg
Study informations
165 subnetworks in total page | file
2310 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 6423
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 | Fisher Score |
|---|
| IPC | 0.7558 | 3.398e-03 | 5.480e-04 | 7.657e-01 | 1.426e-06 |
|---|
| Loi | 0.2297 | 8.123e-02 | 1.161e-02 | 4.305e-01 | 4.062e-04 |
|---|
| Schmidt | 0.6727 | 0.000e+00 | 0.000e+00 | 4.572e-02 | 0.000e+00 |
|---|
| VanDeVijver | 0.7905 | 0.000e+00 | 0.000e+00 | 3.499e-03 | 0.000e+00 |
|---|
| Wang | 0.2608 | 2.790e-03 | 4.483e-02 | 4.010e-01 | 5.016e-05 |
|---|
Expression data for subnetwork 6423 in each dataset
IPC |
Loi |
Schmidt |
VanDeVijver |
Wang |
Subnetwork structure for each dataset
- IPC
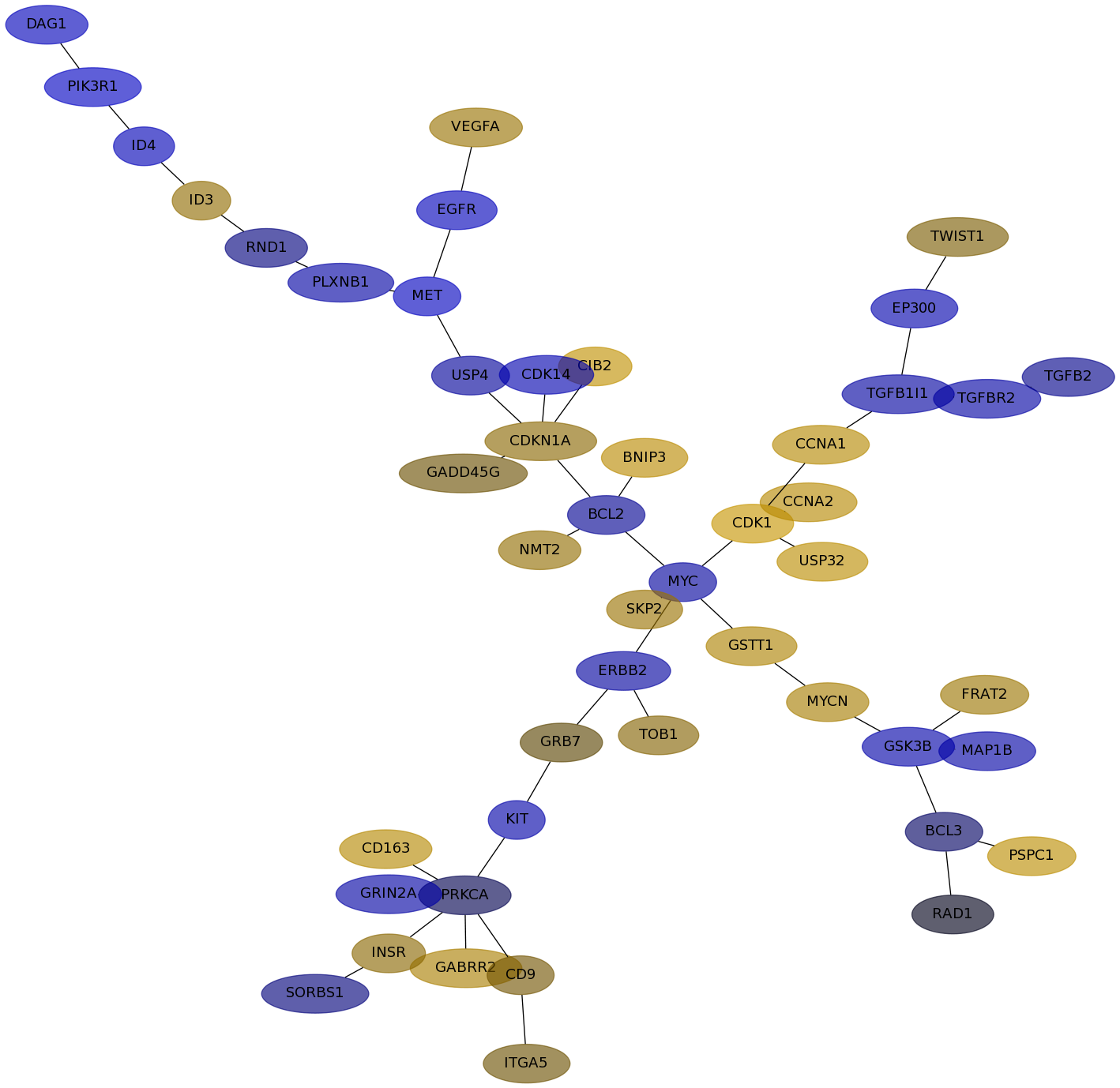
Score for each gene in subnetwork 6423 in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
IPC | Loi | Schmidt | VanDeVijver | Wang |
|---|
| EP300 |   | 113 | 4 | 1 | 4 | -0.149 | 0.087 | 0.102 | 0.052 | 0.025 |
|---|
| USP4 |   | 7 | 256 | 1 | 47 | -0.093 | 0.147 | 0.097 | -0.392 | 0.054 |
|---|
| SORBS1 |   | 57 | 19 | 1 | 13 | -0.043 | 0.015 | 0.314 | -0.072 | 0.200 |
|---|
| GADD45G |   | 43 | 25 | 141 | 134 | 0.030 | -0.029 | -0.207 | 0.201 | -0.055 |
|---|
| CIB2 |   | 18 | 86 | 590 | 564 | 0.228 | -0.149 | 0.106 | 0.127 | 0.190 |
|---|
| GSTT1 |   | 15 | 111 | 1 | 32 | 0.148 | 0.107 | -0.190 | 0.022 | -0.064 |
|---|
| ERBB2 |   | 141 | 1 | 1 | 1 | -0.103 | 0.092 | -0.090 | 0.267 | 0.046 |
|---|
| ID4 |   | 35 | 34 | 236 | 223 | -0.179 | 0.137 | 0.241 | undef | 0.036 |
|---|
| GABRR2 |   | 3 | 557 | 882 | 871 | 0.135 | -0.008 | 0.204 | 0.225 | 0.115 |
|---|
| CDKN1A |   | 110 | 5 | 1 | 5 | 0.064 | 0.254 | 0.030 | undef | 0.082 |
|---|
| TGFB2 |   | 7 | 256 | 525 | 515 | -0.063 | 0.244 | 0.226 | 0.079 | 0.015 |
|---|
| PRKCA |   | 63 | 15 | 1 | 10 | -0.016 | -0.088 | 0.174 | 0.095 | 0.005 |
|---|
| TGFBR2 |   | 7 | 256 | 525 | 515 | -0.115 | -0.028 | 0.136 | 0.105 | -0.190 |
|---|
| PLXNB1 |   | 4 | 440 | 1047 | 1032 | -0.117 | 0.312 | 0.006 | -0.172 | -0.146 |
|---|
| RND1 |   | 6 | 301 | 236 | 239 | -0.046 | 0.085 | 0.022 | 0.082 | 0.113 |
|---|
| CDK14 |   | 7 | 256 | 179 | 201 | -0.151 | 0.011 | -0.071 | 0.212 | 0.107 |
|---|
| CCNA2 |   | 21 | 73 | 179 | 183 | 0.176 | -0.053 | 0.188 | -0.078 | 0.246 |
|---|
| MYC |   | 72 | 10 | 1 | 8 | -0.101 | -0.169 | 0.188 | undef | -0.134 |
|---|
| CD9 |   | 13 | 124 | 525 | 503 | 0.036 | -0.115 | -0.109 | 0.281 | -0.147 |
|---|
| BCL2 |   | 37 | 32 | 83 | 82 | -0.077 | -0.035 | 0.128 | -0.106 | 0.043 |
|---|
| BNIP3 |   | 11 | 148 | 236 | 232 | 0.193 | 0.071 | 0.102 | 0.119 | 0.285 |
|---|
| TWIST1 |   | 50 | 20 | 1 | 14 | 0.044 | 0.194 | 0.061 | undef | 0.219 |
|---|
| MYCN |   | 7 | 256 | 764 | 756 | 0.125 | 0.057 | -0.003 | -0.081 | -0.084 |
|---|
| BCL3 |   | 1 | 1195 | 2178 | 2184 | -0.025 | 0.175 | -0.150 | -0.124 | -0.048 |
|---|
| GRIN2A |   | 12 | 140 | 179 | 192 | -0.113 | -0.068 | 0.162 | 0.125 | undef |
|---|
| MET |   | 6 | 301 | 1047 | 1020 | -0.219 | 0.086 | 0.183 | -0.025 | 0.170 |
|---|
| CD163 |   | 2 | 743 | 1413 | 1410 | 0.175 | -0.206 | -0.019 | -0.063 | 0.038 |
|---|
| KIT |   | 39 | 29 | 1 | 18 | -0.130 | -0.091 | 0.235 | -0.046 | -0.059 |
|---|
| RAD1 |   | 2 | 743 | 1274 | 1260 | -0.003 | -0.168 | 0.120 | undef | 0.001 |
|---|
| PSPC1 |   | 6 | 301 | 991 | 964 | 0.203 | -0.289 | 0.298 | 0.148 | -0.107 |
|---|
| PIK3R1 |   | 127 | 2 | 1 | 2 | -0.237 | 0.381 | 0.065 | undef | -0.002 |
|---|
| SKP2 |   | 29 | 45 | 1 | 23 | 0.092 | -0.043 | 0.237 | undef | -0.009 |
|---|
| ITGA5 |   | 26 | 52 | 179 | 177 | 0.031 | 0.192 | 0.125 | 0.133 | 0.202 |
|---|
| FRAT2 |   | 1 | 1195 | 2178 | 2184 | 0.097 | -0.042 | 0.249 | undef | 0.048 |
|---|
| VEGFA |   | 68 | 14 | 83 | 78 | 0.090 | 0.200 | 0.030 | 0.095 | 0.148 |
|---|
| DAG1 |   | 2 | 743 | 1644 | 1630 | -0.197 | 0.258 | 0.049 | -0.076 | 0.039 |
|---|
| CDK1 |   | 63 | 15 | 83 | 79 | 0.265 | -0.195 | 0.106 | 0.088 | 0.031 |
|---|
| EGFR |   | 117 | 3 | 1 | 3 | -0.195 | 0.240 | -0.205 | -0.187 | 0.065 |
|---|
| NMT2 |   | 7 | 256 | 906 | 876 | 0.077 | -0.296 | 0.134 | -0.124 | 0.217 |
|---|
| MAP1B |   | 69 | 12 | 1 | 9 | -0.123 | 0.263 | -0.107 | 0.304 | 0.100 |
|---|
| ID3 |   | 9 | 196 | 658 | 637 | 0.072 | -0.006 | 0.192 | -0.023 | 0.057 |
|---|
| INSR |   | 96 | 6 | 236 | 221 | 0.063 | 0.173 | 0.135 | -0.014 | -0.072 |
|---|
| GRB7 |   | 58 | 18 | 1 | 12 | 0.021 | 0.115 | -0.072 | 0.229 | 0.129 |
|---|
| CCNA1 |   | 33 | 37 | 412 | 393 | 0.174 | 0.092 | 0.078 | 0.055 | -0.098 |
|---|
| USP32 |   | 2 | 743 | 842 | 854 | 0.204 | 0.263 | 0.137 | undef | 0.062 |
|---|
| GSK3B |   | 79 | 8 | 179 | 169 | -0.130 | 0.292 | -0.129 | 0.291 | 0.057 |
|---|
| TOB1 |   | 6 | 301 | 525 | 517 | 0.055 | 0.097 | -0.069 | 0.107 | -0.081 |
|---|
| TGFB1I1 |   | 49 | 22 | 1 | 16 | -0.105 | 0.229 | 0.143 | 0.195 | 0.298 |
|---|
GO Enrichment output for subnetwork 6423 in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 1.054E-08 | 2.576E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.209E-08 | 1.477E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 1.642E-08 | 1.337E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 5.052E-08 | 3.086E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 6.401E-08 | 3.128E-05 |
|---|
| cell junction assembly | GO:0034329 |  | 6.939E-08 | 2.825E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 1.011E-07 | 3.529E-05 |
|---|
| cell junction organization | GO:0034330 |  | 1.751E-07 | 5.347E-05 |
|---|
| neutrophil chemotaxis | GO:0030593 |  | 3.037E-07 | 8.244E-05 |
|---|
| response to hypoxia | GO:0001666 |  | 3.799E-07 | 9.281E-05 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 3.894E-07 | 8.649E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 2.407E-08 | 5.79E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.759E-08 | 3.319E-05 |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 2.875E-08 | 2.306E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.032E-07 | 6.205E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 1.119E-07 | 5.385E-05 |
|---|
| cell junction assembly | GO:0034329 |  | 1.165E-07 | 4.673E-05 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.084E-07 | 7.163E-05 |
|---|
| cell junction organization | GO:0034330 |  | 3.025E-07 | 9.097E-05 |
|---|
| neutrophil chemotaxis | GO:0030593 |  | 5.299E-07 | 1.417E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 5.98E-07 | 1.439E-04 |
|---|
| negative regulation of survival gene product expression | GO:0008634 |  | 5.98E-07 | 1.308E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.733E-08 | 1.089E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.604E-08 | 6.445E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.79E-08 | 6.739E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.205E-07 | 1.268E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.511E-07 | 1.155E-04 |
|---|
| cell junction assembly | GO:0034329 |  | 3.058E-07 | 1.172E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 4.434E-07 | 1.457E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 5.397E-07 | 1.552E-04 |
|---|
| cell junction organization | GO:0034330 |  | 7.235E-07 | 1.849E-04 |
|---|
| neutrophil chemotaxis | GO:0030593 |  | 1.192E-06 | 2.741E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.354E-06 | 2.832E-04 |
|---|
VanDeVijver file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 1.063E-08 | 1.959E-05 |
|---|
| cell junction assembly | GO:0034329 |  | 1.287E-08 | 1.186E-05 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 1.605E-08 | 9.858E-06 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 2.66E-08 | 1.225E-05 |
|---|
| cell junction organization | GO:0034330 |  | 5.381E-08 | 1.983E-05 |
|---|
| regulation of membrane potential | GO:0042391 |  | 7.451E-08 | 2.289E-05 |
|---|
| cell-substrate junction assembly | GO:0007044 |  | 1.022E-07 | 2.692E-05 |
|---|
| positive regulation of protein kinase activity | GO:0045860 |  | 1.525E-07 | 3.512E-05 |
|---|
| response to hypoxia | GO:0001666 |  | 1.782E-07 | 3.649E-05 |
|---|
| positive regulation of transferase activity | GO:0051347 |  | 1.789E-07 | 3.297E-05 |
|---|
| response to oxygen levels | GO:0070482 |  | 1.998E-07 | 3.347E-05 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| induction of positive chemotaxis | GO:0050930 |  | 4.733E-08 | 1.089E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.604E-08 | 6.445E-05 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.79E-08 | 6.739E-05 |
|---|
| regulation of positive chemotaxis | GO:0050926 |  | 2.205E-07 | 1.268E-04 |
|---|
| positive regulation of cell migration | GO:0030335 |  | 2.511E-07 | 1.155E-04 |
|---|
| cell junction assembly | GO:0034329 |  | 3.058E-07 | 1.172E-04 |
|---|
| positive regulation of cell motion | GO:0051272 |  | 4.434E-07 | 1.457E-04 |
|---|
| peptidyl-amino acid modification | GO:0018193 |  | 5.397E-07 | 1.552E-04 |
|---|
| cell junction organization | GO:0034330 |  | 7.235E-07 | 1.849E-04 |
|---|
| neutrophil chemotaxis | GO:0030593 |  | 1.192E-06 | 2.741E-04 |
|---|
| regulation of mitochondrial membrane permeability | GO:0046902 |  | 1.354E-06 | 2.832E-04 |
|---|



